Endy:Dedicated systems/Translation
Overview
Brink orthogonal ribosomes
RBS design
The first RBS I designed was BBa_B0036. This part uses the RBS described by Brink et al.. The spacing between the RBS and the start codon was set to be the same as that of the consensus E. coli RBS sequence.
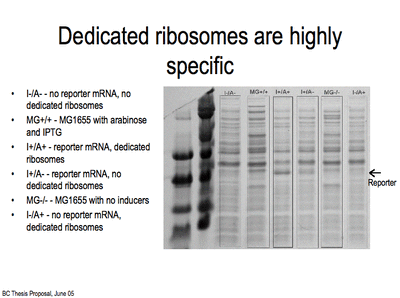
The RiPS from BBa_B0036 are not as high as I would like. Hence I am designing a new RBS. For this new RBS, I will use the spacing between RBS and start codon used by Brink and coworkers. I have obtained high levels of translation using this spacing as shown in the protein gel below using the reporter construct pSD-rbsK2 (obtained from Chris Hayes)
To increase the spacing, I am using the same 5bp sequence as used by Brink et al. This sequence doesn't appear to form a restriction site (it is the first 5bp of an XbaI site) or an E. coli RBS.
Chin orthogonal ribosomes
RBS Design
Registry page for chin RBSs
rRNA Construction
Testing a new RBS-ribosome pair
I'm going to construct a new mutant 16srRNA. I'm going to try two of the sequences published by Chin and Rackham[1]. I'm picking two of the mutants that don't require me to mutate the distant bases that the authors also mutate.
Orthogonal ribosome construction
I'm going to make this via 'Round-the-horn site-directed mutagenesis. This seems like the quickest way to make the full 5 base mutation in one go.
Primer design
pCH1497-ASD1 5'-CacaCTTAccttaaagaagcgtactttgtagtgctcacacag attgtctgatagaaagtga-3'
4rRNA-f primer 5'-TTGTGGTAccttaaagaagcgtactttgtagtgctcacacag-3' Tm = 61.8C
10rRNA-f primer 5'-TGGGATTAccttaaagaagcgtactttgtagtgctcacacag-3' Tm = 61.9C
4/10rRNA-r primer 5'-TGATCCAACCGCAGGTTCCCCTAC-3' Tm = 62.8C
Experimental details
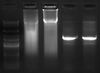
Lane 2 - 4rRNA, phusion
Lane 3 - 10rRNA, phusion
Lane 4 - 4rRNA, pfu ultra
Lane 5 - 10rRNA, pfu ultra
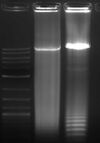
Lane1 - 10kb ladder
Lane 2 - 4rRNA, phusion
Lane 3 - 10rRNA, phusion

Lane 1 - 10kb ladder
Lanes 2-7 - 6 colonies of pCH1497->4rRNA
Lane 8 - positive control PCR on pCH1497
Lane 9 - positive control PCR on I7101
Lane 10 - 10kb ladder
Lane 11-16 - 6 colonies of pCH1497->10rRNA
Lane 17 - negative control, no template

- My plasmid is 10kB so Sean Moore suggested adding fresh polymerase in half-way through. I didn't actually manage to do that on the first attempt.
- Sean also suggested dropping the melting temperature by a few degrees and also the extension temperature. I did 91C for melting and 69C for extension.
- I used Pfu Ultra and Phusion as my polymerases.
- According to the gel I ran of the PCR product (I ran 10μl), phusion worked(?) but pfu ultra did not. Not sure yet why the phusion lanes are smeary. I think I'll redo this and try to add in the fresh polymerase this time.
- Not sure why the PCR product of 10rRNA looks slightly longer than 4rRNA. Maybe just because there is more DNA?
- I did a 10μl ligation rather than the 5 that Sean recommends.
- Got ~50 colonies on each plate of BL21(DE3) with a wide range of sizes(?). I PCR'd 3 big and 3 small on each plate. Only got the right band from the small colonies, not sure why.
- Will start cultures of 4-6 and 10-4, 10-5 and 10-6.
- Sequencing says that 4-6 worked, although it mutated an A to a G just upstream of the ASD. I'll test this anyway to see if it works. I should also check to see if this mutation is due to an error in my reverse primer as it would have been in this region.
- I got no sequence results for 10-6 and 10-5 seems to have had the ASD cut out. I'll try re-prepping 10-6 and see how that sequences.
- From the second gel, start cultures of lanes 3, 6, 11 and 12.
- Colonies 10-1 and 4-2 sequenced correctly. nice.
- In addition, I'm keeping 10-5 from the first sequencing since it has the ASD deleted and 4-5 from the second sequencing as it has a slightly different ASD than expected.
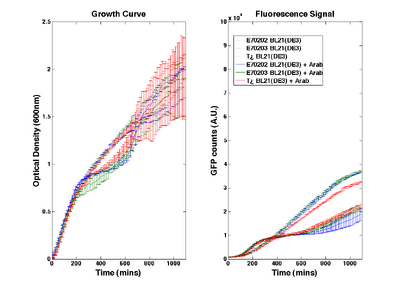
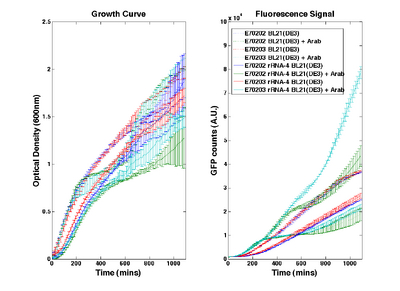
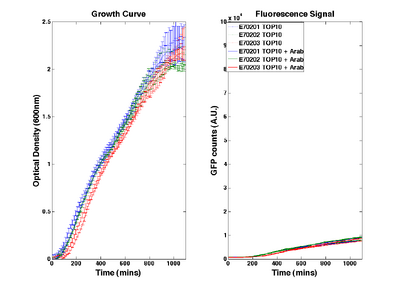
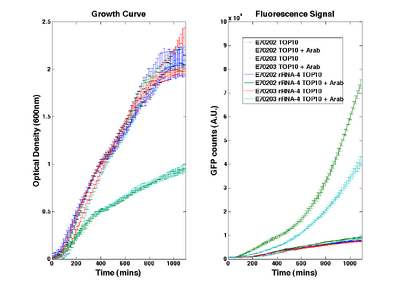
O-ribosome performance