BioMicroCenter:Element Sequencing

Element Sequencing
The Center currently hosts an AVITI platform by Element Biosciences. Element's AVITI platform uses rolling circle amplification with dye-labelled avidites, and is capable of producing more than 1 billion reads per run. The AVITI has 4 independent lanes run across a total of 2 flow cells, with the exception of 2x300 paired-end run kits, which can only run using a single lane per flow cell. Element also supports the conversion of Illumina libraries through their Adept Workflow, adapting Element flow cell binding sequences to Illumina libraries. Index hopping rates are virtually nil, limiting the need for UDI's.
Workflow
Library prep options for DNA or RNA are available for Element sequencing. Once Element-ready or Illumina libraries have been submitted, we will verify fragment size via AATI's Fragment Analyzer or Femto Pulse, adapt Element flow cell binding sites SP5/SP27 if needed, and the quantify the final loading pool via qPCR. We can only guarantee minimum reads per lane if: a)the BMC has performed quality control, b)the samples are high-complexity, especially for the first 5 nt of read 1, and c)submitted libraries are at least 2 nM.
Requirements/Things to Consider
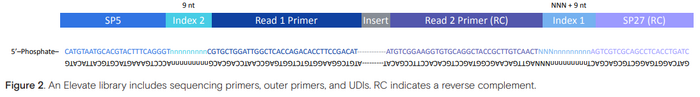
- First 5 cycles of Read 1 must be highly diverse (relatively equal numbers of A, G, C, T per cycle).
- Element ready linear libraries (without Illumina p5/p7) must have 5'-phosphate off of the SP5 sequence for the circularization on the AVITI.
- Library preparations involving truncations, biotin-modifications, poly-A tailing, and/or bead-based normalization will not sequence well, if at all.
- Read direction will be just like MiSeq. The index reads are sequenced first, starting with the sp27 index (which will be read off the sequencing primer site), followed by the SP5 index (which will be read off the SP5 flow cell binding sequence). The read sequences are then read starting with read 1 (using the Read 1 primer sequence), followed by the amplification of the of the reverse strand. Read 2 is then read using the reverse complement of the Read 2 primer sequence.
- Element supports the conversion of Illumina libraries by using the p5 and p7 regions to anneal a stint oligo, followed by the ligation of the SP5 and SP27 flow cell binding sequences. This requires an large input of material (0.5 pmols, or about 20 nM in 30uL) as the ligation process is between 10 and 20 percent efficient. Illumina libraries may also be converted by amplifying on the the SP5 and SP27 flow cell binding sequences using p5 and p7 respectively. Due to the potential for PCR bias, we generally opt for the ligation based conversion method.
Data Handling
Custom Sequencing
Service Element Sequencing SUBMISSION Illumina OR Element libraries MIN VOLUME 15 uL MIN CONCENTRATION 2 nM INCLUDED SERVICES - Quality Control:Fragment Analyzer and qPCR
- Element Sequencing
- Demultiplexing
ADDITIONAL SERVICES - Quality Control
- Element library preperation
DATA FORMATS - FASTQ (stored 1 year)
- BCL (stored 30d)
QUALITY CONTROL - FASTQC
- Basic run metrics (alignment rate, complexity)
- Basic RNAseq metrics (where applicable)
- Basic paired end metrics (where applicable)
- Contamination checks
PRICING LINK SUBMISSION - MIT - ilabs
The G4 Platform
