Beauchamp:CreateAFNIBRIK
Beauchamp Lab wiki
|
|
Previous step is Getting raw data from the scanner Next step is Cortical Surface models overview For the BCM Siemens ScannersThe preferred solution is to use Flywheel system when you are acquiring data on the scanner. This automatically uploads all data from the scanner and converts it to NIFTI (.nii) files. If you have used the Flywheel system, simply log in and download the NIFTI files for your experiment. See this page for instructions: http://www.openwetware.org/wiki/CAMRI:Flywheel Otherwise, on the experiment sheet, you should have written down how many different scan series were collected and what each scan series consisted of. Alternately, you can use the ls command. ls *ima OR ls *IMA The first three digits show the scan series number. The highest number is the last scan series. e.g. 002-000114-152629.ima 003-000048-152134.ima 003-000165-154108.ima 004-000099-153921.ima 005-000040-154821.ima 005-000157-154821.ima 006-000082-160430.ima Means there were six scan series. To see how many images are in each scan series use the following commands (replacing the number "6" with how many scan series you collected): foreach s (`count -digits 3 1 6`)
echo -n $s " "
ls {$s}*ima | wc
end
This will output something like 001 3 3 66 002 183 183 4026 003 183 183 4026 004 176 176 3872 005 192 192 4224 006 192 192 4224 Scan series 001 is a localizer (only 3 images). From the image number it is ambiguous what the other scan series are (anatomical functional). You can use to3d to examine a single file to see if it is an EPI: to3d 003-000165-154108.ima e.g. EPI image with multiple slices, or an anatomy to3d 005-000040-154821.ima One high-resolution anatomical image. Then, to3d can be run accordingly: foreach s (004 005 006)
to3d -prefix series{$s} -session $sess -skip_outliers -anat {$s}*ima
end
for anatomies and foreach s (002 003)
to3d -prefix series{$s} -session $sess -skip_outliers -epan -time:zt 33 183 2000 alt+z {$s}*ima
end
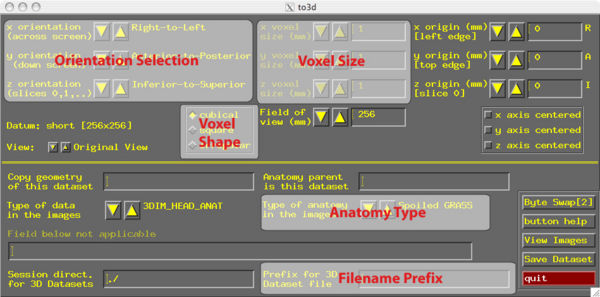
for EPIs. This is the older method for using to3d with Philips dataTo process an anatomical dataset, to3d is quite simple because there is only one timepoint. set session = /Volumes/data1/UT/CD/afni to3d -session $session -prefix CDanat IM_* This creates an AFNI BRIK/HEAD named CD anat from the anatomical DICOM images in the current directory and places them in the "session" directory. AFNI reads information about the images from the DICOM header so that the voxel size and image origin is automatically correct in the BRIK/HEAD file. To process a functional dataset, to3d if more complicated because we must tell it how many timepoints there were (this information is NOT in the DICOM header). Here is the to3d command to create a BRIK/HEAD from a functional run of DICOM images. to3d -session {$session} -skip_outliers -epan -prefix CDr1 -time:tz 110 33 2750 alt+z IM_* 00000001/IM_* 00000002/IM_*
Alternately, we can run to3d on the PAR/REC file. to3d -skip_outliers -epan -time:tz 60 33 2750 alt+z 3D:0:0:80:80:1980:tms_10_10_1.REC Where "1980" is the product of the number of time points and the number of slices (60*33). This requires you to manually enter all of the information about the dataset (voxel dimensions, etc.) in the GUI, as described in the next section. Alternately, a parent dataset can be specified that was collected with the same parameters. to3d -overwrite -prefix test -skip_outliers -epan -geomparent ~/fMRI_1.nii -time:tz 120 33 2000 alt+z 3D:0:0:80:80:3960:C.B.study_2404_2404_F.REC Using to3d from the command lineIf scanner data is obtained in NiFTI format from the UT Philips scanner, then no conversion is necessary (see above). For all other types of scanner data, the program to3d is used to convert the raw data to AFNI BRIK/HEAD files. The preferred way to run to3d is from the command line, so that all parameters are recorded and the process can be automated. To see the to3d options check the help file: to3d –help | more Using to3d from the GUI interfaceIf all of the necessary arguments to create a BRIK/HEAD file are given from the command line, then to3d will create the BRIK/HEAD file and finish. If some arguments are missing, then to3d will display a GUI. For instance, simply go to the directory containing the raw DICOM files and type to3d * from the correct folder. Below is a picture of the GUI interface with the most commonly edited options highlighted. Note that the bottom right of the window includes buttons to view the images, save the dataset, and quit. Normally to3d reads the variable information from the DICOM headers and nothing needs to be changed. Only the filename prefix needs to be set. dcm2niixAs an alternative to using to3d, there is a program developed by Chris Rorden called dcm2niix (the sequel to dcm2nii). This program will sort a folder of dicom images and convert them to all to niftis. One additional nice feature of this program is that it will output the bvals and bvec files for DTI data, including correction for oblique acquisition. Using dcm2niix is pretty straightforward. One generally starts with all of their dicoms in one directory (say, a directory that was just pulled from flywheel or mrisrv). Make a directory where you would like your nii files to be written, and then decide on the filename format. There are other options one can perfuse, but this is sufficient for most purposes. In the example below, my dicoms are in the directory "raw," and I'd like the niftis to be in the directory "niis." I want the filename to be <protocol_name>_<series_number>. mkdir niis dcm2niix -f %p_%s -o niis raw The result is a folder with filenames like "dMRI_dir98_AP_17.nii." Sometimes, multiple reconstructed files will be produced for a single acquisition, and these will be labeled with a letter (e.g., "localizer_HC7_1b.nii"). This can occur when there are multiple slice groups (as is the case for the localizer), or occasionally for partially-constructed files. In the latter case, there is almost always a file with just the number (for example, "MPRAGE_3.nii," in addition to "MPRAGE_3a.nii," "MPRAGE_3b.nii," etc) and the .nii with just the number is the version you would use going forward. As with all things in imaging, it is prudent to check the files after this step as funny things can happen. Be especially vigilant to check for R-L flips (because a flip in any direction can happen, but it's a lot more noticeable if you happen across a brain that's upside down.) |