BME100 s2016:Group5 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
(http://www.promega.com/resources/protocols/productinformationsheets/g/gotaqcolorl essmastermixm714protocol/)
have the same forward primer and reverse primer
samples will be crosscontaminated
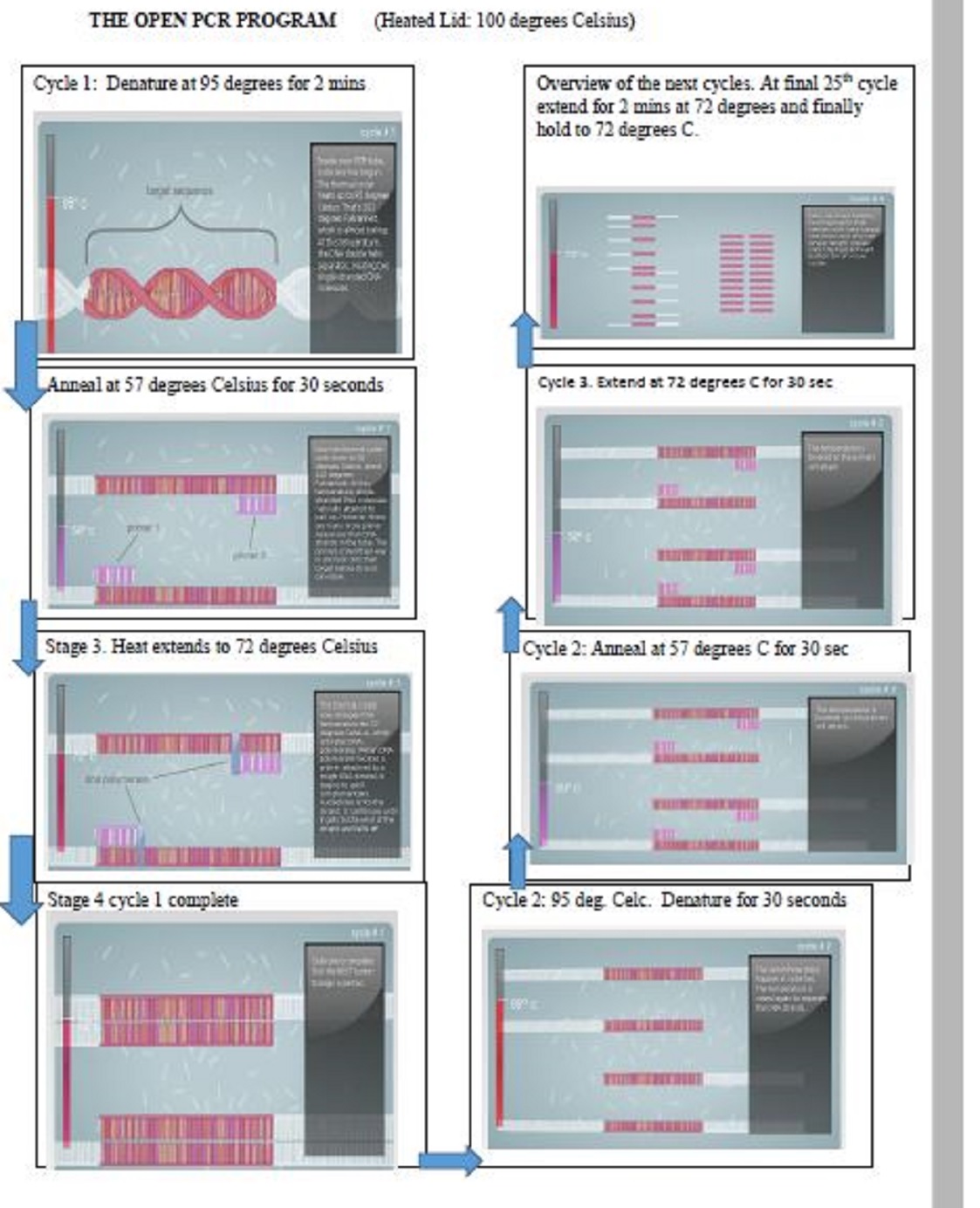
In this lab the OpenPCR Program will be run as follows:
Steps in diagrams are shown below. Pictures Extracted from the PCR vitual lab at: http://learn.genetics.utah.edu/content/labs/pcr/ NB: (typo) the final step should read hold at 4 degrees Celsius and not 72 degrees Celsius. for better viewing see: File:PCR procedure.pdf
Research and DevelopmentQuestion 1 Template DNA: Positive and Negative control Primers: Short pieces of DNA that are made in a laboratory Taq Polymerase:Thermostable polymerase named after the thermophilic bacterium Thermus aquaticus. Most important enzyme for PCR reaction. Deoxyribonucleotides (dNTP’s): A monomer (a single unit of DNA) Each composed of nitrogenous base, deoxyribose sugar, and a phosphate group. It is specific to DNA. Question 2 In the first step (INITIAL STEP) the temperature is raised to 95 degrees Celsius for 3 minutes because the polymerase is activated by heat, in order the reaction to occur. Second step is DENATURE where the components are held at 95 degrees Celsius for 30 seconds. In this step both strands of deoxyribonucleic acid unwinds and separates into single-stranded strands through the breaking of hydrogen bonds and other intermolecular interactions. Third step is called "Anneal" which occur at 57 degrees Celsius for 30 seconds. Now that we have two strands of DNA,primers will complementary attach to these strands to make new stable strands of DNA. After Anneal,the temperature is changed to 72 degrees Celsius for 30 seconds in order the extension process to occur. At this step the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding dNTPs that are complementary to the template. We keep this temperature for 3 minutes more to make sure the extension process is finished. At the end we change the temperature to 4 degrees Celsius to stabilize the components, so they don't start the reverse reaction.
Adenine anneals to Thymine Guanine anneals to Cytosine
Question 4 The steps where base-pairing occurs, is during the second and third step. The second step which is annealing, is when the two primers bind appropriately to their sequence. The mixture is cooled between 45-72 degrees celsius during this step. The third step which is extension, is when the reaction is heated to 72 degrees celsius. This allows for DNA polymerase to act. DNA polymerase extends the primers, which then nucleotides are appropriately added to the primer using target DNA as a template.
PCR - The Underlying Technology
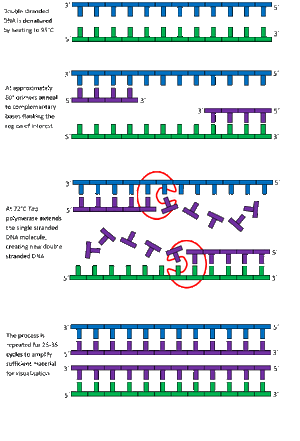 We do not own the image and it can be found to this link: ttps://www2.le.ac.uk/departments/emfpu/genetics/explained/images/PCR-process.gif Note: before "ttps" should be add an " h " so the link would be correct .
SNP Information & Primer DesignBackground: About the Disease SNP A single nucleotide polymorphism(SNP) is a nucleotide(basic structural unit of DNA) that can occur in more than one form. For this project we are focusing on the SNP rs36686. The rs36686 SNP is a single nucleotide polymorphism that is found in homo sapiens. The variation is located on the 19:17811986 chromosome. B3GNT3 is the gene associated with this SNP and the B3GNT3 gene has been linked to non-Hodgkin Lymphoma. The full name for the B3GNT3 gene is UDP-GLcNAc:betaGal beta-1, 3-N-acetylglucosaminyltransferase 3. This gene is responsible for several functions that include carbohydrate metabolic process, cellular protein metabolic process and O-glycan processing. The disease-associated allele for this gene is found in the sequence "CGC" at the location 17811986. Primer Design and Testing To find the non-disease forward primer we recorded a 20 base sequence that ended at the position of the SNP. For the non-disease reverse primer we counted 200 bases ahead of the SNP position, starting with the lower base at that position we took the preceding bases to create another 20 base sequence. We then made disease forward and reverse primers by changing the final base on the forward sequence. After testing the non-disease primers on the UCSC In-Silico PCR web tool (http://genome.ucsc.edu/cgi-bin/hgPcr?command=start), we received a matching 220 base pair sequence from chromosome 19:17811986. The disease primers yielded no matches, this would be due to the variation in the SNP that causes the disease. Images from: http://genome.ucsc.edu/cgi-bin/hgPcr?command=start
| ||||||||||||||||||||||||||||||||||