BME100 s2016:Group1 W1030AM L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||
EKCRNC Technologies
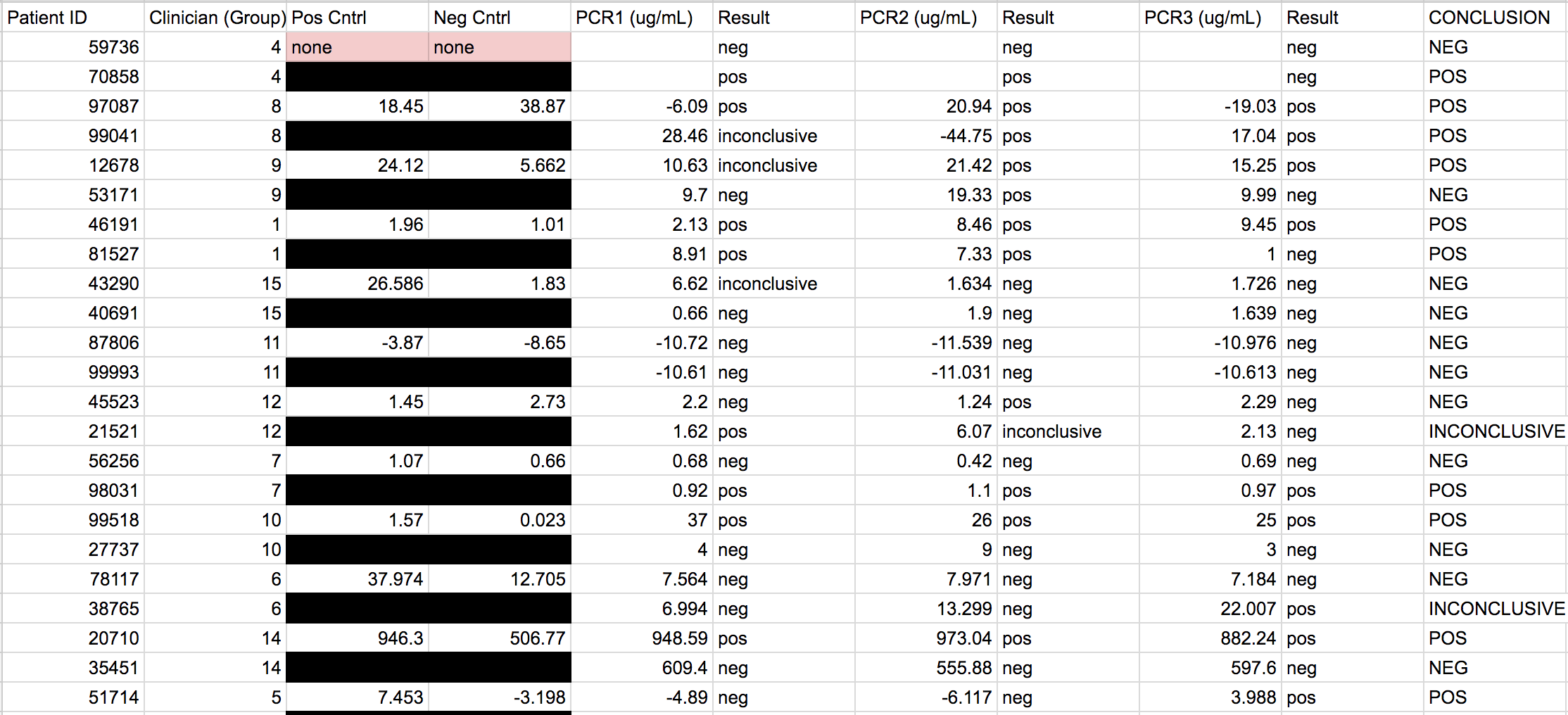
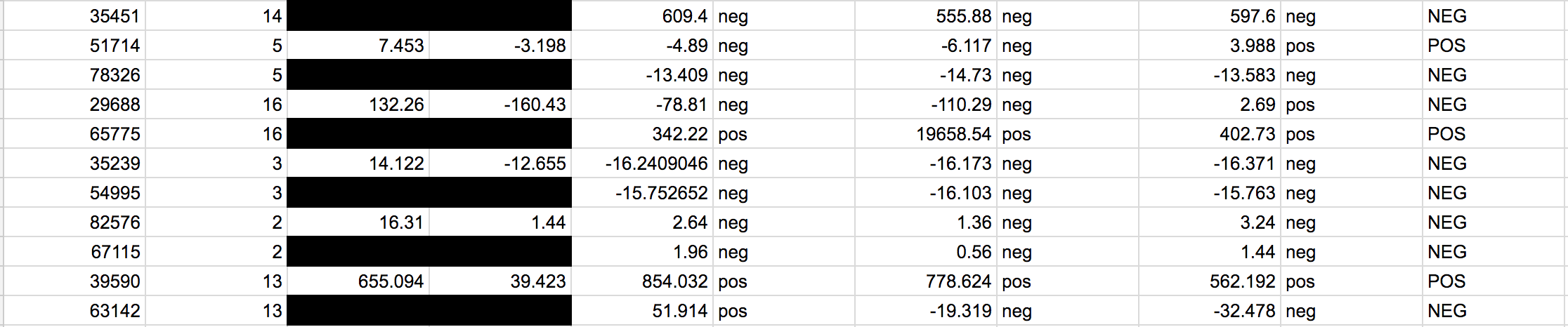
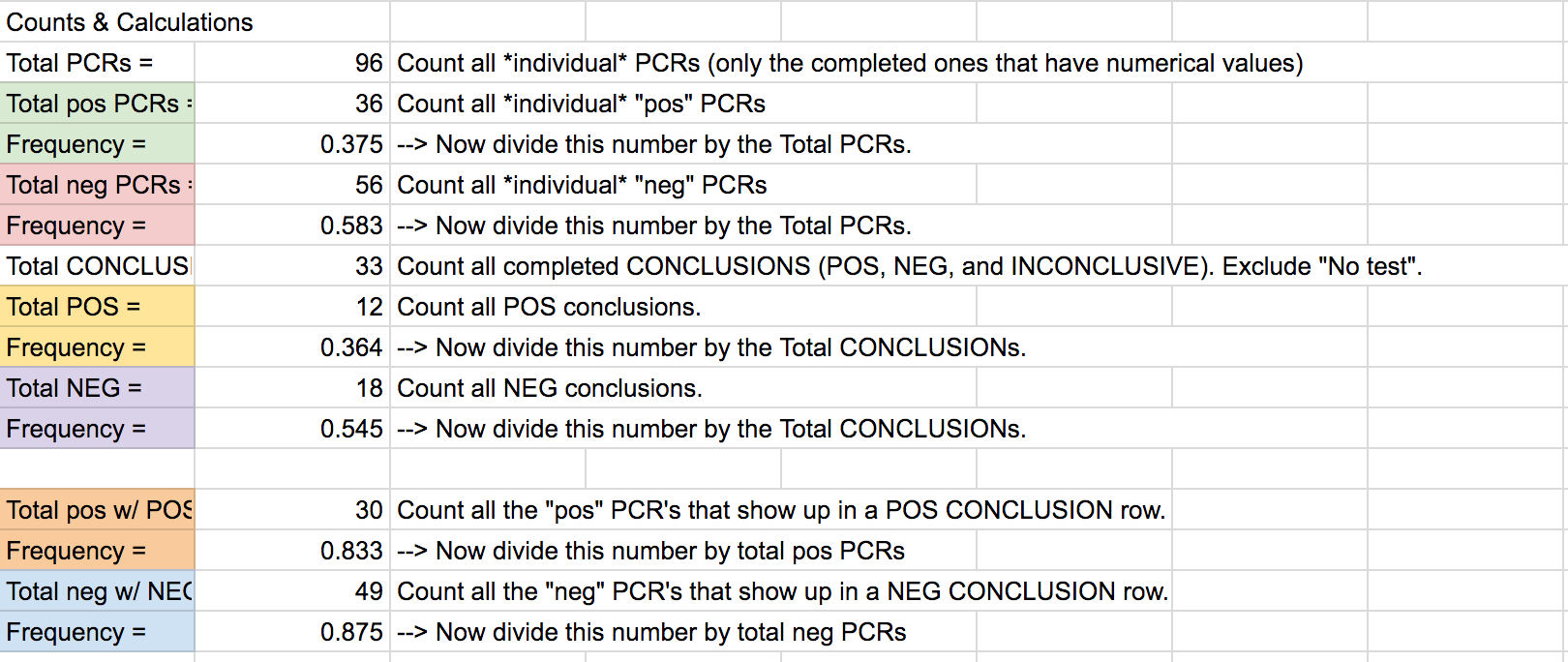
LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System In BME 100, the process of polymerase chain reaction (PCR) was used to test the DNA of patients for the disease-associated SNP. The class was divided into 17 teams with 6 students each. Each group got 2 patients to diagnose leading to a total of 34 patients being tested. In order to prevent error or minimize the amount of error three replicates were created for each patient. Along with creation of multiple replicates, controls were put in place for the actual process of PCR and for the calibration in ImageJ. The control for PCR was a test tube which contained DNA from a person who definitely carried the SNP disease whereas the negative control was the DNA of a person who definitely did not carry the SNP disease. The patients tested had not yet been tested for the disease. For the ImageJ analysis of the pictures taken the positive control was 80 microliters of SYBER Green I - which glowed a fluorescent green whereas the negative control was 80 microliters of pure solution that did not glow at all. To make sure each trial was consistent with the previous trials a micropipette was used and a single 80 microliter drop was analyzed. The final data was as follows:
What Bayes Statistics Imply about This Diagnostic Approach
The PCR was less reliable during calculations 3 and 4. There is about 40% reliability of PCR for concluding that the patient will develop the disease given a positive final test conclusion, and about a 70% reliability for concluding that the patient will not develop the disease given a negative final test conclusion.
- The usage of wrong amounts of the solutions. - The lack of trials (only one per solution). - Ambient light from the room may have broken down the SYBER Green I to cause a false negative. Intro to Computer-Aided DesignTinkerCAD Our Design
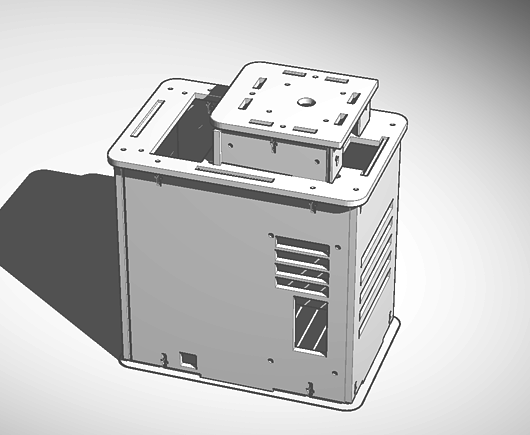
Our design mimics the Open PCR machine's general body (4 walls, bottom, lid) and the inner workings of the machine (heat sink and fan, power supply, circuit board, etc). However, we redesigned our machine to include self-distribution within the machine. Instead of requiring the user to transfer solutions into the PCR tubes through a pipette, the machine will do the work for you. All the user needs to do is insert a container of the solution into the specified slot, and the machine will transfer predetermined amounts of the solution into each of the PCR tubes with efficiency and accuracy. Our final design is also more visually appealing.
Feature 1: ConsumablesIncluded in our packaging we will include PCR mix, primer solution, buffer, and plastic tubes specifically designed for our PCR machine. These are arguably the most important materials to include in a consumable package since they are all specifically designed to mesh easily and fully with the PCR machine designed by our company. Feature 2: Hardware - PCR Machine & FluorimeterThe general body of the Open PCR machine and all of its inner workings (heat sink and fan, power supply, circuit board, etc) will be used in our final PCR machine design. The flourimeter, however, was excluded because our group wants to focus on the polymerase chain reaction itself, and not fluorescence detection. The main problem our group noticed during the PCR lab was that the measurements taken by the pipette were not completely exact due to user error. Transferring solutions also took up the majority of our time during the lab. With these in mind, we redesigned the Open PCR machine to create a PCR machine that transfers solution from a larger container into the individual PCR tubes on its own. This makes the PCR process easier for the user because it eliminates the need for pipette transfers, and the results obtained will be more accurate.
| |||||||