BME100 s2016:Group1 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program
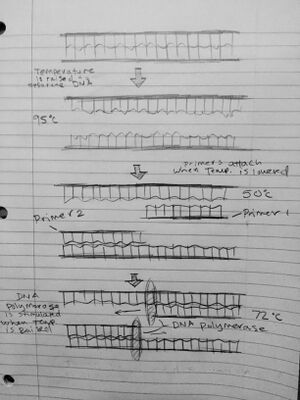
Research and DevelopmentPCR - The Underlying Technology Research and development - the underlying technology PCR works by heating and cooling DNA. Initial DNA is placed into a container where primers are added. Primers bind to specific segments of interest once the DNA is separated in heating. When DNA is heated to the specific heat of 95 degrees celsius, DNA begins to unwind and separate. When the DNA strands become single stranded, the solution is cooled to a temperature of 50 degrees celsius. As the temperature cools, DNA will naturally want to rejoin due to the electrical charges on both strands but before this happens primers interrupt this process through a manner similar to competitive inhibition and manage to bind onto the strands of DNA before they can reform. With primers bound onto the DNA strand the process of DNA replication can begin. The solution is then heated to 72 degrees celsius in order for DNA polymerase to activate. DNA polymerase is an enzyme that binds to specific primer regions and begins adding nucleotides to the strand until it reaches the end. This process is repeated several times until around cycle three when the desired targeted DNA segments are produced. This is a result of DNA strands with both primer one at one end and primer two at one end. This is a result of successive replication of DNA at specific primer points through several cycles. As this process continues with these desired targeted DNA segments also multiplying the results quickly become pronounced and in abundance. One of the most interesting aspects about PCR found in research was the specificity of the DNA polymerase. DNA polymerase is destroyed at high temperatures except for a specific type found in a strain of bacteria called Thermus Aquaticus. This polymerase is referred to as Taq. A side note, Taq is a powerful example of the implications of patenting a DNA strand as its manufacture and ownership has been contested several times in court. Because Taq does not degrade at high temperatures, its usage is incredibly important in PCR as it can survive the changing heat (which has powerful implications on its value).
SNP Information & Primer DesignBackground: About the Disease SNP SNP is a short genetic variation which a specific sequence of DNA differs by a single base pair, from another similar sequence. SNPs are found in all life forms with DNA. This can affect physical appearance, susceptibility to disease, and response to medication of the life form. Once a region of DNA has been selected, a primer can be designed to amplify the DNA. Heat is used to separate the DNA strands. Primer 1 and primer 2 attach to the free stands and are built upon with nucleotides, and the cycle repeats until there are billions of copies of the DNA sequence. To design a primer it must be able to bind with the target DNA. The primer consists of 18-22 base pairs, primer 1, and a reverse sequence called primer 2. The SNP should be located on the end of the primer so that it becomes amplified.
| ||||||||||||||||||||||||||||||||||