BME100 s2016:Group12 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
have the same forward primer and reverse primer
samples will be crosscontaminated
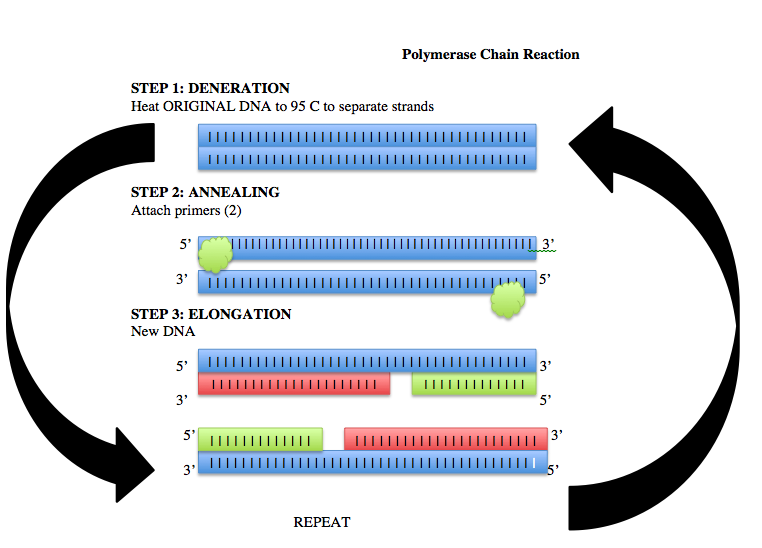
Research and DevelopmentPCR - The Underlying Technology The Different PartsIn order for the entire PCR process to work, there must be several different components. Each component serves its own purpose in the reaction and ultimately plays its own part in the final result. Template DNA is the primary component of this process as it is the source of the target DNA the process was developed for. It provides the original piece of genetic coding which will be singled out and duplicated many times over in the end. Primers are the first component that actually starts the PCR process. The primers indicate where the target DNA is and, therefore, shows what will be copied. They also provide a binding site for the Taq polymerase, allowing them to find the target DNA sequence. Taq Polymerase, once it binds to the site the primers indicate, reads the template DNA strands and binds nucleotides to the target DNA that was isolates by the primers. In binding the nucleotides to the target DNA, the Taq polymerase actually duplicates the target DNA strand. Finally, the Deoxyribonucleotides are the building blocks of DNA which, in this case, serve as the materials with which to put together the duplicated target DNA strand. Bringing it all togetherFor each step of the PCR process which takes place inside of the PCR Machine, a certain temperature associates it. The Initial Step takes place at 95˚C for 3 minutes and is when the strand of Template DNA unwinds in order to allow for separation. Next, while still at 95˚C, the template DNA experiences the Denature step in which it separates into the forward and reverse strands in 30 seconds. Then, as the nucleotides of the DNA are exposed, the primers can attach to the target DNA in the Anneal stage which takes place at 57˚C for 30 seconds. The temperature then rises again to 75˚C, and the Extend stage takes place. With the increased temperature, the Taq polymerase is activated in 30 seconds, and it locates the primer sites on the template DNA. The Final Step then takes place with the temperature steady at 75˚C for 3 minutes, and, in that time, the Taq polymerase binds the deoxyribonucleotides to the target site on the template DNA, thereby duplicating the target DNA. To finish the process, the temperature is brought back down to 4˚C, and the Taq polymerase is then deactivated, leaving the newly formed strands of the target DNA. Base-PairingThroughout the PCR process, there are two steps which experience base-pairing- the Anneal and the Final Step. Anneal pairs the target DNA with the primers, and the Final Step pairs free deoxyribonucleotides with existing target DNA. Especially in the latter, it is important to note the nucleotides will specifically pair with certain other nucleotides. Adenine bonds with Thymine, and Cytosine bonds with Guanine.
SNP Information & Primer DesignBackground: About the Disease SNP Single nucleotide polymorphisms, often referred to SNP represents differences in each nucleotide at a certain position in the genome, this shows a variation between genes amongst people and their likeliness to develop a certain disease. SNPs are found within the DNA between genes and can act as "markers" to identify where the diseased genes are located. When SNPs exist in a gene or near a gene that specific gene's function can be changed or altered. If SNPs are located in a non-coding region they can result in a greater risk of cancer and can alter MRNA structure whereas if they are located in coding regions they can cause a change in the amino acid sequence of a protein.
| ||||||||||||||||||||||||||||||||||
Lab B
Nucleotide - a compound consisting of a nucleoside linked to a phosphate group. Nucleotides form the basic structural unit of nucleic acids such as DNA
Polymorphism - the presence of genetic variation within a population, upon which natural selection can operate.
What species is this variation found in? – Homo Sapiens or Humans
What chromosome is the variation located on? - Chromosome: 19:17811986
What is listed as the Clinical significance of this SNP? - None
Which gene(s) is this SNP associated with? - B3GNT3
What disease is linked to this SNP? - Immune function may predispose to the development of non-Hodgkin lymphoma.
Part 2
What does B3GNT3stand for? - B3GNT3 stands for beta-1,3-N-acetylglucosaminyltransferase 3
What is the function of B3GNT3? To find out, click the B3GNT3 link. Under Table of Contents (right side) click Gene ontology. Write the first three unique terms you see - This gene encodes a member of the beta-1,3-N-acetylglucosaminyltransferase family. This enzyme is a type II transmembrane protein and contains a signal anchor that is not cleaved. It prefers the substrates of lacto-N-tetraose and lacto-N-neotetraose, and is involved in the biosynthesis of poly-N-acetyllactosamine chains and the biosynthesis of the backbone structure of dimeric sialyl Lewis a. It plays dominant roles in L-selectin ligand biosynthesis, lymphocyte homing and lymphocyte trafficking. Acetylglucosaminyltransferase, lacto-n-tetraose, poly-N-acetyllactosamine.
What is an allele? - An allele is one of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome.
The disease associated allele contains what sequence? - CAC
The numerical position of the SNPis: - 17,811,986
Part 3
Nondisease forward primer (20 nt): - GCAACGCTACCTCGGGCGTGC
The numerical position exactly 200 bases to the right of the disease SNPis: - 17,812,186
Nondisease reverse primer (20 nt) - GGAGGAAGGTGTCGCCCCTT
Part 3: 14.
Disease forward primer (20 nt) - GTGCGGGCTCCATCGCAACA
Disease reverse primer (20 nt) - GGAGGAAGGTGTCGCCCCTT