BME100 s2015:Group8 9amL4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program
Initial Step: 95°C for 2 minutes Number of Cycles: 35 Denature at 95°C for 30 seconds Anneal at 57°C for 30 seconds Extend at 72°C for 30 seconds Final Step: 72°C for 2 minutes Final Hold: 4°C
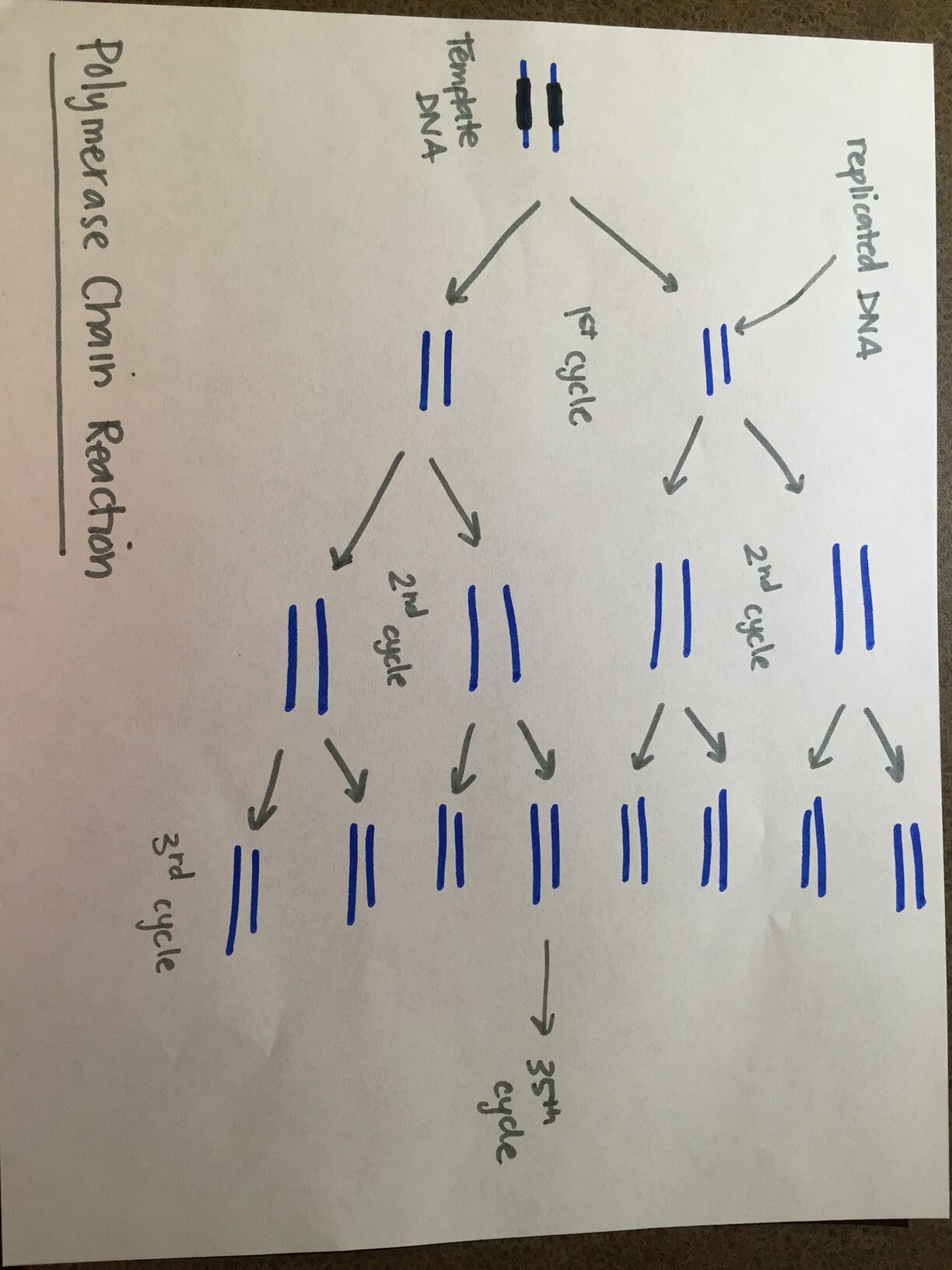
Research and DevelopmentPCR - The Underlying Technology The following are the functions of assorted portions of a PCR reaction: the template DNA is the original DNA in which a specific section or sequence is targeted. The primers are essentially the beginning and end nucleotides of the targeted sequence. The Taq Polymerase attaches to a single strand of DNA, reads each nucleotide, and then attaches the appropriate base pairs. The deoxyribonucleotides are the base pairs that Taq polymerase attaches to the newly created single strand of DNA in order to complete the strand. The purpose of heating the DNA to 95 degrees Celsius for three minutes is to ensure that the DNA is unwinding from its usual double-helix shape and is ready for the denaturing step of PCR. Next, when the temperature is maintained at 95 degrees Celsius for an additional thirty seconds, the denaturing process actually occurs: the two strands of DNA completely unwind and split. During the subsequent step, the “annealing”, where the temperature is decreased to 57 degrees Celsius for thirty seconds, the two strands naturally have a proclivity for rejoining. The “Extend” step allows for the Taq polymerase to take action, joining the appropriate base pairs and amplifying a specific sequence with the assistance of primers. In the FINAL STEP, the isolated segments of DNA are allowed to solidify, and return to their natural state (double-helix formation). Lastly, the FINAL HOLD portion allows for the sections of DNA to be cooled adequately for storage. The base pairing is as follows: Adenosine binds to Thymine, while Cytosine binds to Guanine. The process of base pairing begins in the “annealing” step. The temperature at this step allows for base pairs to begin to come together naturally. Base pairs, which exist in a lock-and-key format, ensure that only the appropriate base pairs bind to the isolated strand. The temperature of the “extend” step, 72 degrees Celsius, is when the Taq polymerase is binding base pairs at its fastest rate.
| |||||||||||||||||||||||||||||||||