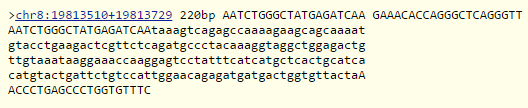BME100 s2015:Group5 12pmL5
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 5 WRITE-UPProcedureSmart Phone Camera Settings
Placing Samples onto the Fluorimeter
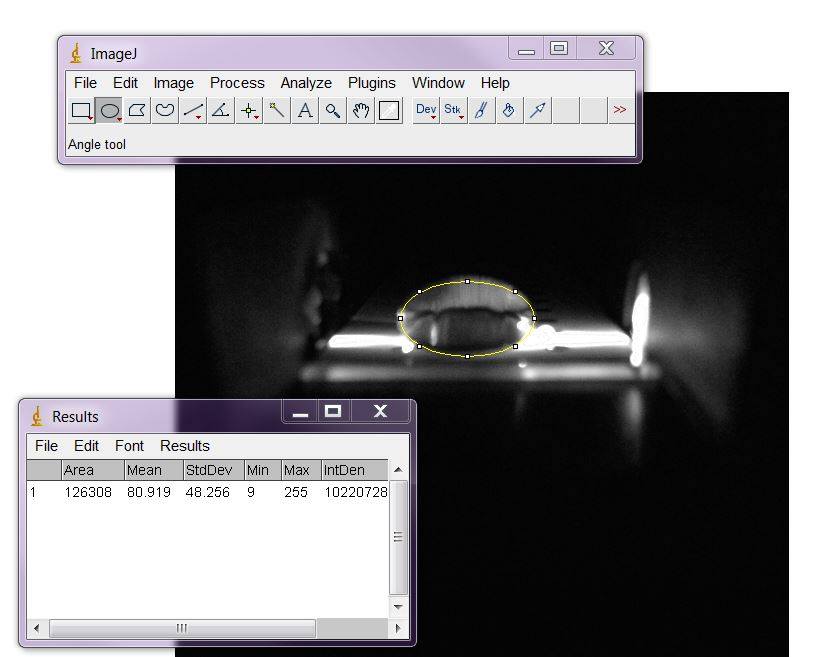
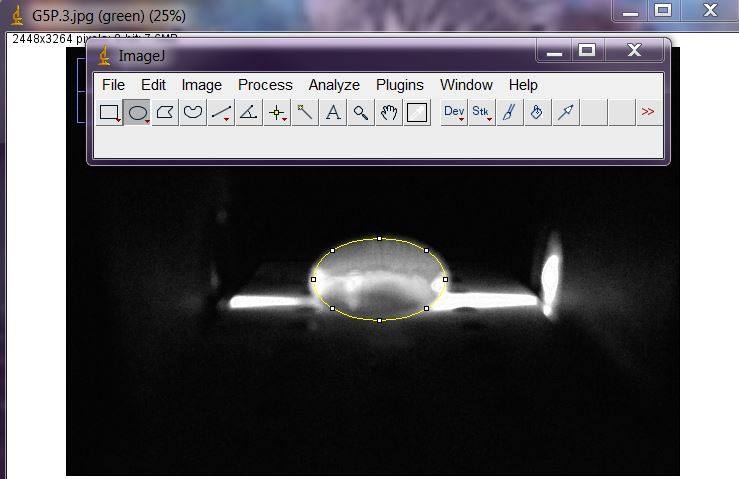
Data AnalysisRepresentative Images of Negative and Positive Samples
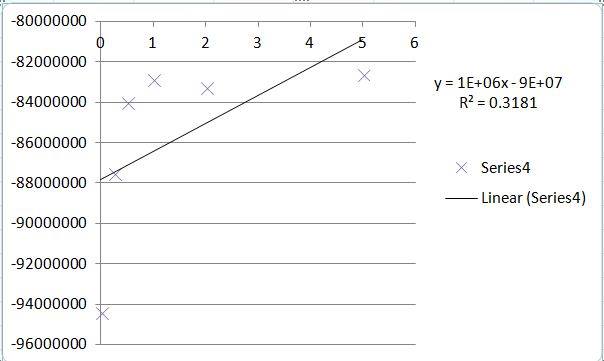
PCR Results Summary
Observed results
Conclusions
SNP Information & Primer DesignBackground: About the Disease SNP The language of DNA is similar to the language in which people write in. Much like written languages, errors can occur as a result of the misspelling of a word. The language of DNA is not written in letters or numbers, but instead in molecules called, "nucleotide bases," which are composed of a nitrogenous base, a pentose (a five-carbon sugar), and at least one phosphate group, and are found in DNA as one of four bases: Adenine, Thymine, Guanine, and Cytosine (A,T,C, and G). The equivalent of typos in genetics are found in SNPs. SNPs, or Single Nucleotide Polymorphisms, are variations in a genome at the base-pair level, where during replication, a single base pair can be copied incorrectly during DNA replication. This leads to the base-pair being substituted, added to, or replaced in a way that is not a correct copy of the original genome, which can lead to millions of outcomes. In fact, it is believed that the 10 million SNPs in the human genome are what account for the differences between humans. Among other things, SNPs can lead to differences in appearance (e.g. hair color or skin tone), responses to medical treatments (e.g. allergies or resistances), and even susceptibility to disease (e.g. hereditary predispositions to diseases like cancer). The specific consequence of an SNP in the genome that is being examined in this experiment is the "disease SNP." The "disease SNP" is actually a missense SNP (where the correct nucleotide is replaced by an incorrect allele) in the species Homo Sapien, or humans, with pathogenic significance. Alleles are alternative forms of a gene that are created via mutations and can be found in the same place in a chromosome. The disease SNP is found in one such allele, which normally contains a sequence of the bases Adenine- Adenine--Thymine (AAT). The disease-causing allele, however, features an SNP in the second adenine base, changing the sequence to Adenine-Guanine-Thymine (AGT), and is found at position 19956018. A direct consequence of this SNP is the condition of type I hyperlipoproteinemia (a form of Hyperlipidemia), which stems from an overabundance of cholesterol in the body, and is hereditary. The link between the missense SNP in question and type I hyperlipoproteinemia is that the allele that the "disease SNP" affects is responsible for the production of lipoprotein lipase (LPL). LPL finds its role in the body as a homodimer (a quaternary structural protein) in the heart, muscle, and adipose tissue, but more importantly, it is responsible for the breaking down of triglycerides (triglyceride hydrolase) and as a ligand/bridging factor for the complex process of lipoprotein uptake. Because of this integral role that LPL plays, any mutation (or SNP) that affects the production of LPL is cause for extreme medical concern and immediate research.
Every PCR reaction requires two primers. The first primer is a twenty-base long sequence, ending in the correct nucleotide of the affected allele (A). This sequence is 5'AATCTGGGCTATGAGATCAA. The second primer, the reverse primer, is found in 200 bases forward from the affected "A" SNP, and is 5'GAAACACCAGGGCTCAGGGTT. Upon sequencing these two primers in an online sequencing service, it was determined that the sequence was 220 base-pairs (bp) long, meaning that the two primers are valid, as the first primer is set 20bp before the affected allele, and the second is set 200bp after. Because the two match up, and allow for a successful sequencing, the two primers are valid and correct, and ready to be used. The results of the sequencing of the two primers is shown below: Naturally, because there must be an experimental group, the primers must also be prepared for the diseased allele. All that must be done for this is the modification of the final "A" in the forward sequence, as that base is the affected base. The primer sequence, thus, is changed to 5'AATCTGGGCTATGAGATCAT. The reverse sequence 200bp from the allele is unaffected, as a switching of the base doesn't add or subtract length from the entire genome, meaning that the reverse sequence remains 5'GAAACACCAGGGCTCAGGGTT. Upon attempting to sequence these two primers, an error is encountered, as the first sequence is not a valid sequence in the human genome when matched with the second sequence, as the SNP is a mutation. The result of the attempted sequence is shown below: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||