|
OUR TEAM
LAB 1 WRITE-UP
Initial Machine Testing
The Original Design

The Open PCR (picture shown above) is a machine that uses heating and cooling cycles to multiply DNA strands. The DNA heats to 95 degrees Celsius, separating the double helix structure of the DNA. Then, it cools down to 57 degrees Celsius to attach the primers to the beginning of the desired strand. The final step of the cycle reheats the DNA to 72 degrees Celsius. This attaches the DNA polymerase, an enzyme which attaches the complimentary bases to the template DNA strand, to the primer, replicating the DNA. After 35 cycles of replicating, there is an abundance of the desired strand of DNA, outnumbering the initial DNA.
Experimenting With the Connections
When we unplugged (part 3) from (part 6), the machine screen did not turn on. (This wiring was responsible for the connection between the screen and the circuit board.)
When we unplugged the white wire that connects (part 6) to (part 2), the temperature sensor did not work. It read -40 degrees Celsius rather than 24 degrees Celsius. This leads to the observation that the white wire must be what allows an accurate temperature reading to be displayed.
Test Run
The Open PCR machine was tested for the first time on October, 23, 2013 at approximately 10:15 a.m. The circuitry of the machine is fairly complex, however the various parts of the machine were defined and comprehended before the experimentation began. The Open PCR machine ran at an average rate of about two hours a test or three and a half minutes a cycle, with accurate cycling temperatures.
Protocols
Thermal Cycler Program
- Initial denaturation step: one cycle at 95°C for 3 minutes.
- Denaturation step: 35 cycles at 95°C for 30 seconds.
- Anneal step: one cycle at 57°C for 30 seconds.
- Extended step: one cycle at 72°C for 30 seconds.
- Final extension step: one cycle at 72°C for 3 minutes.
- Final holding: refrigeration of DNA at 4°C.
DNA Sample Set-up
| Positive Control: cancer DNA template Tube label: PC |
Patient 1 ID: 46398 Replicate 1 Tube label: P11 |
Patient 1 ID: 46398 Replicate 2 Tube label: P12 |
Patient 1 ID: 46398 Replicate 3 Tube label: P13
|
| Negative Control: non-cancer DNA template Tube label: NC |
Patient 2 ID: 73945 Replicate 1 Tube label: P21 |
Patient 2 ID: 73945 Replicate 2 Tube label: P22 |
Patient 2 ID: 73945 Replicate 3 Tube label: P23
|
DNA Sample Set-up Procedure
- Step 1 : Gather supplies and label test tubes (as shown above with the table).
- Step 2 : Add Primer 1 to each PCR test tube, which attaches to either end of the DNA at hand.
- Step 3 : Add Primer 2 to each PCR test tube, attaching to the second site of the DNA strand.
- Step 4 : Add Nucleotides to each PCR test tube.
- Step 5 : Add DNA Polymerase to each test tube.
- Step 6 : Put PCR test tubes into thermocycler. Collect data.
- Step 7 : Store test tubes for later use.
PCR Reaction Mix
- 8 PCR reaction mix tubes with 50 μL each.
- Each tube contains Taq DNA polymerase, MgCl2, and dNTP's.
DNA / Primer mix
- 8 DNA / Primer mix tubes with 50 μL each.
- Each contains a different template DNA.
- All tubes will have the same forward primer and reverse primer.
Research and Development
PCR - The Underlying Technology
Polymerase Chain Reaction (or PCR) is a procedure used to create copies of DNA.
DNA Base-Pairing
Deoxyribonucleic acid (DNA) is the genetic material of living things. It has a double helix structure, similar to a spiraling ladder with two sturdy sides and rungs running up the middle. At each "rung," there are two bases attached to the spines. Four kinds of bases can be found in DNA: adenine (A), cytosine (C), guanine (G), and thymine (T). Each base can only pair with one other base, however. Adenine only pairs with thymine and cytosine only pairs with guanine. A single base, combined with the sugar and phosphate group in the spine, is called a deoxyribonucleotide (dNTP).
Components of PCR
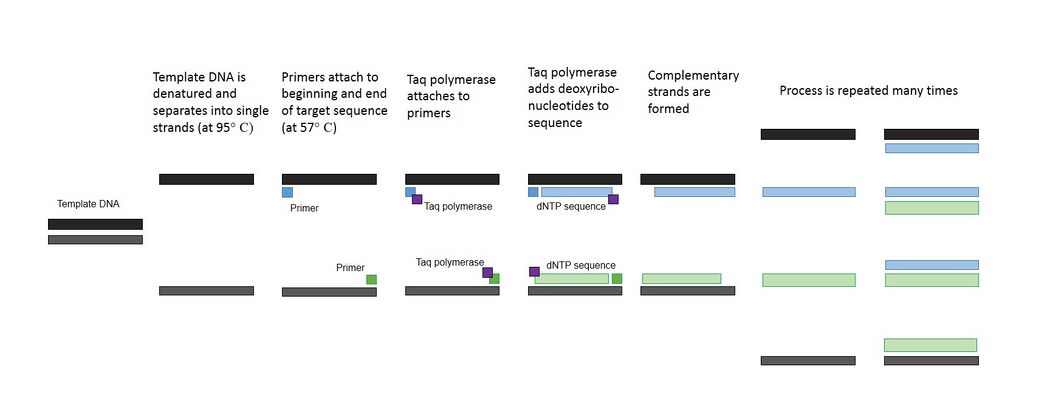
PCR is composed of multiple steps, each essential to the overall process. The procedure starts with template DNA, which are the target strands that will be denatured and amplified. Primers are added to the template DNA to attach to specific sites on the separated DNA strands. Two primers are used - one to mark the beginning of the target sequence and one to mark the end. An enzyme called Taq polymerase will then attach onto the primers and begin adding dNTPs down the sequence, creating a complementary DNA strand. Magnesium chloride (MgCl2) catalyzes the Taq polymerase and helps stabilize the DNA.
Steps of PCR
Step 1: 95°C for 3 Minutes (Initial Step)
- The mixture is heated to 95°C to begin activating the PCR components. This also causes the template DNA to begin to denature. It will unravel and separate into two strands, with two distinct sequences of unpaired bases.
Step 2: Denature at 95°C for 30 Seconds
- The DNA continues to denature from its double helix structure into single strands.
Step 3: Anneal at 57°C for 30 Seconds
- The machine cools, which allows the two primers to attach to the DNA sites at the beginning and end of the target sequence.
Step 4: Extend at 72°C for 30 Seconds
- The machine is heated, activating the Taq polymerase. The enzyme attaches to the primers, and the magnesium chloride allows it to begin attaching complementary dNTPs to the exposed bases. This creates a complementary strand that completes the single original strand of denatured template DNA.
- Steps 2-4 can now be repeated to double the amount of new strands created. The new double helix DNA is denatured, annealed, and extended into more double helixes, which are in turn denatured, annealed, and extended. These steps can be repeated as many times as necessary, although usually 25-40 cycles are run to create over a billion copies of the target DNA sequence.
Step 5: 72°C for 3 Minutes (Final Step)
- Taq polymerase creates complementary strands for all remaining single strands.
Step 6: 4°C (Final Hold)
- The reactions are stopped.
PCR Process
Sources:
|






