BME100 f2013:W1200 Group13 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||
OUR COMPANY
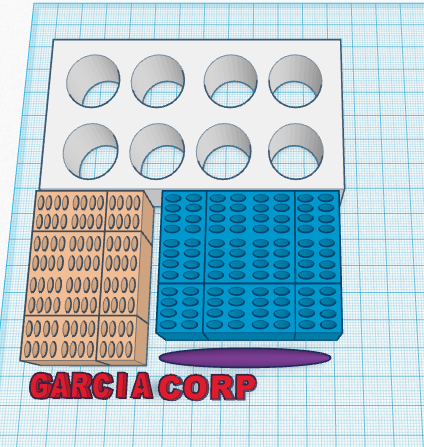
LAB 6 WRITE-UPComputer-Aided DesignTinkerCAD Our group used TinkerCAD to create new PCR tubes. The design of the tubes was created while considering organization and ease of handling. The openPCR machine that we used in class is set up to hold four rows of four. We set up our tubes to be in two groups, Each group connected for handling and organization. The first group is the + control and the three trials of patient 1, thus our tubes are labeled so "+" "1-1" "1-2" "1-3" are visible and have a slight grade so they can be felt as well without getting stuck in the machine. The same was done for the second group of tubes with the - control and patient 2's trials respectively.
TinkerCAD could be used practically for this experiment because the design for the OpenPCR machine could be redesigned. The machines that we used had several issues. These issues revolved around overheating and user construction flaws. With these flaws in mind the way we would redesign the machine would be to print the shell of the machine a little bigger, then repositioning the fan so it has two parts, one blowing cold air in and the other moving hot air out. On top of that the heat sink should be moved next to the hot air outtake fan so it is constantly cooled and displacing heat. This will help the overheating issue. The next thing could be to print the machine with small latches or pins because the screws made a screw driver necessary and also cause issues with missing pieces and splitting wood. With each printed perhaps with an L hook and a button the machines would all be put together identically, thus eliminating the user construction problem.
Feature 1: Cancer SNP-Specific PrimersBackground on the cancer-associated mutation
Primer design
How the primers work: Primers are custom pieces of DNA that are made to attach at the top strand of DNA and also at the end of the specific segment of DNA. The two strands are not identical, but they are complimentary. For this experiment the primers are designed to attach to the target strand of DNA then once the solution is placed in the OpenPCR machine it replicates that targets strand and after only a few cycles its already replicating exponential amounts of the target strand. This gives us more of a sample to test from and helps eliminate any factors that we don't want. The primers essentially let us pick a target strand of DNA and isolate it for replication.
Feature 2: Consumables Kit
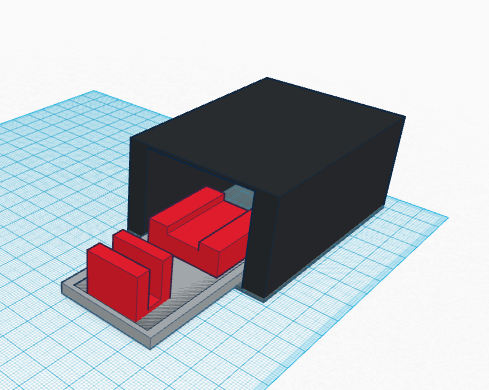
Feature 3: PCR Machine HardwareThe PCR machine will be packaged separately from the box of consumables that way it does not damage any of the items. They can be bought together in a complete package but they will not be stored together to avoid any damage to the PCR machine or the consumables. The tube holders in the PCR machine would have a stopper in the corner to make sure that the lid is on all the way and not breaking the tubes. This is a simple modification that can create more accuracy. The stopper would be the slightly higher than the tubes but not enough to make the tubes not touch the lid. It is higher enough to take the pressure from the lid off the tubes, but not enough to effect the experiment.
Feature 4: Fluorimeter Hardware
Bonus Opportunity: What Bayesian Stats Imply About The BME100 Diagnostic ApproachThe Bayesian statistics equation is written as P(A|B)=(P(B|A)*P(A))/P(B). Bayesian statistics is a way to determine the reliability of predicting cancer and detecting cancer SNPs. Calculation 3 asked what the probability is that the patient will develop cancer given they have the cancer DNA sequence. The P(A|B) value for calculation is less than one. Calculation 4, P(A|B) was also less than one, making both of the calculations very inaccurate. Calculation 4 asked what the probability is that the patient won’t develop cancer given a non-cancer DNA sequence. Therefore, the reliability of CHEK2 PCR for predicting cancer isn't accurate and shouldn’t be relied for cancer detection. There is human error from the PCR machines and also some of the data was not used because results couldn’t be drawn from them due to incomplete tests. Since the number of tests was smaller than expected, this could have affected the outcomes. If both the P(A|B) and P(B|A) value were closer to 1 the test would be a reliable way to determine if a patient has cancer or not. However since they are very small it is not a reliable determination for the prediction of cancer in a patient. |
||||||