BME100 f2013:W1200 Group10 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||
|
OUR TEAMLAB 1 WRITE-UPInitial Machine TestingThe image above is a picture of the device OpenPCR. OpenPCR is an open source thermocycler that copies DNA molecules in a compact machine using the common reaction PCR, which stands for Polymerase Chain Reaction. PCR involves amplifies the replication of DNA strands and specifically replicating certain segments of the genome. The OpenPCR machine uses the enzyme polymerase to create a complementary strand of DNA, once the two original strands of DNA are split through temperature increase. The product is then two identical strands of DNA, which the process can be done multiple times to make thousands of copies. OpenPCR uses a system of heating and cooling components to activate the reaction and to stabilize the newly made DNA strands between cycles. PCR is important because it is used commonly for detecting viral infections, food safety testing, manipulating DNA in genetic engineering, and much more.
When we unplugged (part 3) from (part 6), the machine's power to the LED screen was turned off. When we unplugged the white wire that connects (part 6) to (part 2), the sensors on the heat plate were disconnected. They were registering the temperature at -40 degrees Celsius until plugged back in.
We first tested the OpenPCR machine on October 23rd. We opened up the device to figure out how the inside was all connected. To make sure that the machine could run through the trials correctly, we inserted empty test tubes and ran a fake experiment. We entered the temperatures and number of cycles that we wanted, but didn't add any DNA to the test tubes, this way we could just test the machine to make sure it was running properly. At the end of 25 trials, our machine was operating correctly and it was ready to use for the actual PCR experiment.
ProtocolsThermal Cycler Program Heated Lid: 100*C Initial Hold: 95*C, 3 minutes 35 Cycles: 95*C, 30 seconds -> 57*C, 30 seconds -> 72*C, 30 seconds. Final Step: 72*C, 180 seconds. Final Hold: 4*C, forever. DNA Sample Set-up
1.) Thaw master mix. 2.) On ice prepare 12.5µl master mix, .25-2.5µl 10µM upstream primer, .25-2.5µl 10µM downstream primer, 1-5µl DNA template, and 25µl Nuclease-Free Water. 3.) adjust volumes to fit required reaction volume. PCR Reaction Mix Master mix contains: Reaction buffer (pH 8.5), Taq DNA polymerase, 400µM dATP, 400µM dGTP, 400µM dCTP, 400µM dTTP and 3mM MgCl2.
All primer mixes contain the same forward and reverse primers.
Research and DevelopmentPCR - The Underlying Technology
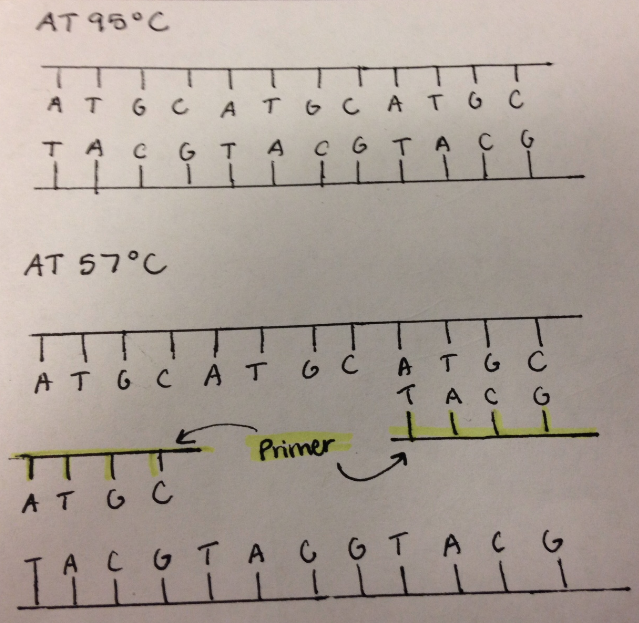
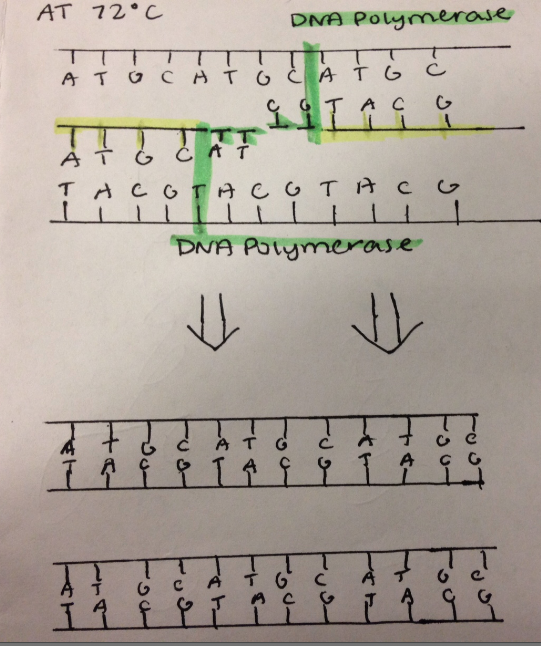
In the PCR reaction, the template DNA strand serves as the template of DNA which is to be copied by various enzymes. Enzymes read the template stand and compose a complementary strand based on the sequence of the template DNA. RNA Primase adds primers which are short strand of RNA to the DNA strands in order for DNA Polymerase to attach dNTPS as DNA Polymerase can only add nucleotides to an existing strand. Primers indicate the position where copying must begin on the DNA strand. The addition of primers also prevents the two DNA strands from rejoining. Taq Polymerase is a protein complex that copies DNA by adding dNTP's to the position on the strand as indicated by the primers. In order for DNA Polymerase to add dNTP's to the the DNA strand, magnesium chloride is necessary to catalyze the enzyme. Deoxyribonucleotides are unpaired nucleotides consisting of a phosphate, deoxyribose sugar, and a nitrogenous base that are used by DNA Polymerase to create the new complementary strand.
The Polymerase Chain Reaction goes through thermal cycling to copy DNA. First the temperature is raised to 95°C for 30 seconds. At this temperature denaturing of DNA occurs. The hydrogen bonds holding the nucleotides together break apart, and the the double helix of DNA is effectively split in half. Next, the temperature is lowered to 57°C for another 30 seconds. This is the temperature at which DNA naturally attempts to rejoin. Instead, specific sequences of nucleotide are attached to by the lab made primers. This prevents the DNA from rejoining, and indicates to the polymerase where to begin copying. In the final step of the cycle, the temperature is then increased to 72°C. At this temperature, the DNA polymerase begins to attach the excess dNTPs to the complementary pairing on the DNA. This process is catalyzed by the magnesium chloride in the solution. After 3 cycles, the desired dna sequence begins to form itself. These are indicated by strands of DNA with primers on both ends. The number of desired strands increases exponentially with each further cycle. After 30 cycles, there are 1 billion target strands. The solution is now effectively a pure solution of the target sequence. | |||||||||