BISC 219/F10: RNAi Lab 10
Series 3 Reverse Genetics: Scoring RNAi Worms and RNA Isolation
3 days before next lab:
- Come into lab and find your stack of plates.
- On 2 of the experimental plates add 2 L4 wild type (N2) hermaphrodites
- On 2 of the experimental plates add 2 L4 rrf-3 hermaphrodites
- On 1 of the control plates add 2 L4 wild type (N2) hermaphrodites
- On 1 of the control plates add 2 L4 rrf-3 hermaphrodites
- Wrap all of your plates in an elastic and stick in your lab day box in the worm incubator set at 23°C
Phenotypic Analysis
Today in lab you will examine your RNAi worms and their progeny.
- Examine worms with a known bli-1 mutation to see what knockdown looks like.
- Compare your RNAi worms to worms that have a known bli-1 mutation- how do they compare?
- Examine your control worms to review wild type phenotype in comparison to mutants
- Compare your RNAi worms to the control worms - are they the same phenotype? Different? What do the “control” worms tell you?
- Examine your RNAi’d wild type (N2) and then your RNAi’d rrf-3 worms. Do you see any differences between the two treated strains of worms?
Be sure to record all of your results in your lab notebook.
Take pictures of your control and RNAi worms to use in the results section of your next paper.
RNA Isolation
The theory of RNAi states that the mRNA specfic to the gene of interest will be degraded inside the cells of the worm because dsRNA initiates an interference RNAi pathway and amplification that affects future generations. The degradation of a gene's mRNA transcript will prevent the production of the protein product of that gene and thus, possibly, show downregulation or post-transcriptional silencing of that gene through observation of an aberrant phenotype.
If your RNAi worked exceptionally well, most of your worms on a treated plate will show a visible aberrant phenotype while your control untreated worms of the same strain will not. If 80% or more of the worms on one of your RNAi plates showed some or significant downregulation of bli-1 (seen by aberrant phenotype), collect all of the worms on that plate by washing them off with 1X PBS + Tween 20 buffer. The mutant worms will tend to "stick" to the bacteria on the medium so you will have to squirt PBST onto the plate and swirl it around the plate until you see the worms (and some bacteria) floating in the buffer. Tilt the plate to get the liquid and worms to one side and use a sterile Pasteur pipet to squirt liquid to dislodge them and to transfer the worms and buffer into a 1.5 ml microcentrifuge tube. Repeat for your untreated control of the same strain worms in a separate microcentrifuge tube.
(If less than 80% of your worms showed a visible phenotype change compared to untreated, control worms, consult with your instructor. The best thing to do would be to pick 100 worms displaying the aberrant phenotype and place them into 100 μL of 1X PBS + 0.25% Tween20 buffer in a 1.5 ml microcentrifuge tube. Repeat for your control worms of the same strain in a separate tube. Get the worms off your pick by shaking it in the PBST. Make sure you have gotten the worms off the pick and into the tube! )
Let the worms settle to the bottom of the microfuge tube for approximately 5 minutes. Look for worms in the bottom of the tube (you can use the microscope if you have trouble seeing them but don't). Carefully remove the top portion of buffer with a Pasteur pipet until you are left with settled worms in about 50 to 100 μL of the buffer. Be careful to avoid sucking up the worms!! Wash the worms once more by adding about a ml of PBST, letting the worms settle. The TWEEN dissolves and kills the bacteria and those bacteria should remain in the supernatant. Again CAREFULLY suck out most of the buffer until you are left with about 50-100 μL of worms in buffer at the bottom of your tube.
In order to break through the tough cuticle we must freeze-crack the worms. Place the tubes with the worms and buffer in the tube in the -80°C freezer for 15 minutes. Thaw the worms quickly by wrapping your warm hand around the tube when you remove them from the freezer.
The next steps require the use of Phenol and Chloroform - please wear gloves, a lab coat and eye protection. We will work in the hood! Phenol will burn your skin on contact.
- Add 500 μL Trizol (Invitrogen #15596-026) to both control and treated worms. WORK IN THE HOOD
- Incubate both tubes at room temperature for 15 minutes (dissociates the nucleoprotein complexes)WORK IN THE HOOD
- Add 100 μL of chloroform to each tube then making sure the caps are snapped on tightly. WORK IN THE HOOD
- Shake each tube at the same time, vigorously for 15 seconds.WORK IN THE HOOD
- Incubate the tubes for 2 minutes at room temperature WORK IN THE HOOD
- Spin them at 12,000 rcf in the refrigerated microcentrifuge or in the Eppendorf bench top centrifuge with the appropriate rotor for 15 minutes at 4°C. IN LAB
- After spinning, you will notice a top aqueous phase that contains the RNA while the interphase (cloudy layer in the middle) contains the DNA. The lower red phase contains the proteins. There is about 150 to 200μL of RNA so you must be very careful to transfer ONLY the clear top aqueous phase to a clean 1.5 ml microfuge tube in small aliquots of about 50 μL at a time using a Pasteur pipet. It is much more important NOT to get the DNA interphase than to get every bit of your RNA layer. BE VERY CAREFUL NOT TO GET THE DNA INTERPHASE - IT WILL MESS UP YOUR RESULTS! Repeat for your second tube. WORK IN THE HOOD
- Add 250 μL Isopropanol (NOT ETHANOL) to each RNA isolate solution; mix by inversion a few times. WORK IN THE HOOD for this mixing step.Incubate at room temperature for 10 minutes. This will begin the RNA precipitation. From this incubation on you may work at your bench.
- Spin at 12,000g for 15 minutes at 4°C being careful to align the hinge side of the microcentrifuge tubes facing out (up).
- Remove the supernatant and look for a pellet on the hinge side of each tube - you may or may not see one.
- Wash the pelleted RNA (visible or invisible) very carefully by slowly adding 200μL of 75% Ethanol (NOT ISOPROPANOL) to each tube on the side opposite the pellet. Immediately draw all of the ethanol back off. Before you discard the ethanol in your waste container, check to see if your pellet is still on the side of the tube or if it is floating in your pipet tip. If the later, place it back into the tube and spin the tube again before attempting to remove and discard the wash.
- Turn the tubes upside down on a Kimwipe™ and let them air dry for 5 minutes.
- Add 20 μL of DEPC treated water.
DNase Treatment
To be sure that the only nucleic acid you will amplify in your RT PCR reaction is RNA and not contaminating DNA we will treat our samples with an enzyme DNase. We will use TURBO DNase from Applied Biosystems. It is a genetically modified enzyme that is more efficient form of DNaseI than the natural enzyme.
- Measure the RNA concentration using the NanoDrop2000 in the equipment room. See the instructions below.
- If your concentration is >200 ng/μL, dilute some of your sample in DEPC treated water so that you have a 20 μL sample that is 200 ng/μL. Check with your instructor when you have made your calculations of how to dilute your sample. If your concentration is less than 200 ng/μL, go on to the next step without dilution.
- Add 2 μL of 10X Turbo DNase buffer so that the final concentration of buffer is 1X in each RNA isolate solution.
- Add 1 μL of TURBO DNase (2 units) to each sample.
- Incubate at 37°C for 30 minutes.
- Add 2 μL of 150 mM EDTA to your sample. What is the final concentration of EDTA?
- Incubate at 75°C for 10 minutes to heat kill the DNase.
- Determine your nucleic acid concentrations again using the NanoDropper. Record ALL information in your notebook. You will need the concentrations for next week.
- Give your DNA free RNA isolates to your instructor to freeze until next week.
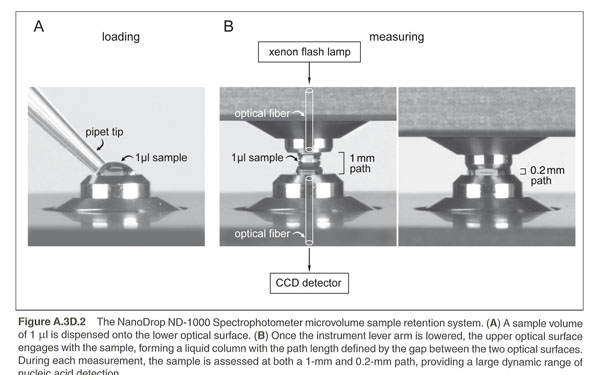
Measuring Nucleic Acids with a Nanodropper
There is a Nanodroppers in the BISC Equipment room, L308. The ThermoScientific NanoDrop 2000 measures DNA or RNA by taking Absorbance at a particular peak absorbance wavelength. These spectrophotometers use only 1 microliter of sample and do not require cuvettes. The sample is held in place by fiber optic technology and surface tension that holds the sample in place between two optical surfaces that define the pathlength vertically and dynamically. Measurement can be assessed in a range of 2 to 3700nm/microliter dsDNA. These are expensive machines so make sure you follow the directions carefully and ask your instructor for guidance as needed.
More information is available from the manufacturer's website at: | http://www.nanodrop.com/HowItWorks.aspx
Using the Nanodroppers
1. Clean the upper and lower optical surfaces of the sample retension device by pipetting 2 microliters of clean deionized water onto the lower optical surface. Close the lever arm and tap it a few times to bathe the upper optical surface. Lift the lever arm and wipe off both optical surfaces with a Kimwipe.

2. Open the NanoDrop software from the Desktop of the computer and select the appropriate nucleic acids module (RNA).
3. Initialize the machine by placing 1μL of clean deionized water onto the lower optic surface, lower the lever arm, and select initialize from the NanoDrop software. Once initialization is complete (~10sec.), clean both optical surfaces with a Kimwipe.

4. Perform a blank measurement by loading 1μL of deionized water and select Blank. Often the blank will be something other than deionized water if you have your sample in a different solvent.
Note that as in traditional spectroscopy, the blank will be subtracted from subsequent measurements. If you want to determine the contribution of a specific buffer or diluent, measure the buffer or diluent first using distilled water as a blank. If the buffer does not contribute to the Absorbance reading, then deionized water will be fine to use as the blank. Water or buffer should always be measured to be sure that the instrument has been zeroed properly. The measurement of water or buffer/diluent should be zero or very close. All measurements are automatically normalized to the appropriate wavelength.
5. Measure the nucleic acid sample by loading 1μL of sample and selecting "measure". (Make sure that you have selected RNA rather than DNA for measurement) Record your RNA concentration. Once the measurement is complete. Clean both optical surfaces with a Kimwipe and the machine is ready for the next sample.
You should ensure that the appropriate constant (50 for dsDNA or 40 for RNA) has been chosen. The software automatically calculates the nucleic acid concentration. If the calculation is done by hand, the A260nm is represented as a 1cm path for convenience, even though 1-nm and 0.2nm paths are actually used during the measurement cycle.
Clean Up
When the last sample was been measured, clean the sampling device by repeating step 1.
Outline of Experimental Design for REVERSE Genetics Project
Where are you now in this process?(What have you done so far; What's next?)
Make the feeder strain of bacteria
- Amplify gene of interest by pcr ;
- Restriction Enzyme digestion of amplified DNA to create "sticky ends" for ligation;
- Clean up DNA (remove enzymes);
- Cloning: ligate gene into vector plasmid with amp resistance gene ;
- Transform competent bacterial cells;
- Select for transformants on media with ampicillin;
- Perform colony pcr on several transformants to be sure to find one colony containing a vector plasmid with the gene of interst
- Culture the selected colony from colony pcr to create a lot of copies of these bacteria
- Isolate the cloned plasmid DNA from that cultured colony by miniprep;
- Retransform isolated plasmids (with gene interest) into HT115 (DE3)cells genetically modified to have impaired ability to degrade RNA;
- Select for transformants on media with ampicillin
- Choose an isolated colony to culture and make lots of feeder strain bacteria;
- Induce expression of C. elegans gene dsRNA from the pL4440 vector in the bacteria by IPTG induction.
- Seed NM lite worm growth media plates with feeder strain produced as described
Plate wild type C. elegans worms (N2 and rrf-3 strains) on feeder plates made as described (containing bacteria expressing dsRNA of our gene of interest).
Observe phenotype change in progeny caused by RNAi silencing or knockdown of the gene of interest compared to control worms of same strains that were NOT fed feeder strain bacteria.
Isolate RNA from RNAi worms and control worms of same strains.
Perform RT-PCR (Reverse Transcriptase)to amplify the C. elegans gene of interest, using worm RNA and then cDNA as template. The RNA is isolated from treated RNAi worms and untreated worms of the same species.
Visualize the worm gene of interst in the pcr product by agarose gel electrophoresis and compare the amount of amplified DNA in RNAi treated vs. untreated worms.
Assignment
Remember to check the Assignment section of the wiki for instructions about the graded assignment due in the next lab and check the Weekly Calendar for other work to accomplish before the next lab.