BISC220/S13: Mod 3 Lab 11
Lab 8: Cell Culture
Lab 9: Apoptosis - DNA
Lab 10: Apoptosis - Protein 1
Lab 12: Imaging Presentations
Media Recipes
Western Blot of PARP-1
Today we will finish the Western Blot using PARP-1 antibody.
Yesterday we blocked your membrane with 10 ml of blocking buffer 5% milk in phosphate buffered saline (1X PBS) with 0.25% Tween (commonly referred to as PBS-T) for 1 hour. We then removed the blocking buffer and added mouse anti-PARP-1 antibody (BD Pharmingen #556362) in 5% milk in phosphate buffered saline with 0.25% Tween (PBST). The anti-PARP-1 was diluted 1:1000 (so you need 10 ul for 10 ml of buffer). These blots were rocked overnight at 4°C. You will begin with the next step. *Note: you will need to save the primary antibody for use by other lab groups.
- Return the primary antibody solution to the conical centrifuge tube and give it back to your instructor (try to recover as much volume as possible). Pour PBST wash buffer onto the nitrocellulose so that it is submerged by approximately 0.5 cm of liquid. Incubate 7 min. on the shaker.
- Pour off the wash buffer into the sink. Add fresh wash buffer and incubate another 7 min. on the shaker.
- Repeat Step 2 once more (a total of 3 7-min. washes).
- Add the HRP-conjugated goat anti-mouse secondary antibody 1:10,000 in 10 ml blocking buffer (i.e. add 1 µl of antibody to 10 ml of blocking buffer). Incubate 45 min. on the shaker.
- Pour off the secondary antibody solution into the sink (you do not need to save this).
- Wash three times with PBST (7 min. each wash) as you did after the primary antibody incubation. After the last wash, remove all residual wash buffer by tilting the blot container onto a paper towel. *Do not proceed with the following steps until your instructor is available to help you with the film exposure and developing. Leave your blot in the final batch of wash buffer until that time. The following directions for performing the ECL may be different depending on which manufacturer’s kit we use. Check with your instructor to see if the following step (ratio of reagents 1 &2) needs to be modified before proceeding.
- Pipet 1 ml of ECL detection reagent 1 into a 15-ml conical tube and add 1 ml of reagent 2. Invert to mix. It is important for the two solutions not to mix until just before you use them! Make sure that you record all the kit information in your lab notebook as the reagents are proprietary.
- Pour the mixed ECL developing solution onto your blot. Make sure all the membrane is in contact with the liquid by rocking the container slightly back and forth by hand for 1 min. Discard the ECL reagents in the sink.
- Working quickly but carefully, use forceps to place your blot onto a piece of paper towel. Using another piece of paper towel, gently blot the nitrocellulose to remove excess liquid.
- Use forceps to place the membrane blot—protein side up—between the two plastic sheets of a page protector.
- With your instructor, take the cassette into the equipment room. Using the new GelDoc we will image your Western Blots. Put your blot in the machine – protein side up. Close the door and open the Imaging software. Click on New Protocol. Choose Blots and then Chemi.
Fluorescence Microscopy
Background on Fluorescence Microscopy
A substance is said to "fluoresce" when it emits light following excitation by light or another energy source. The fluorescence microscope is specially designed to filter light, since fluorescent molecules absorb and emit light at different wavelengths (Figure 1). The light source passes through one filter that allows only the wavelength that is absorbed by the fluorescent molecule of interest to pass through to the sample. This excites the fluorescent dye. A second filter allows only the emitted wavelength to pass through to the eyepiece and thus the eye of the observer.
Today you will view cells from the VP-16 timecourse that have been stained with the DNA binding dye Hoescht-33342. What will you expect to see in the VP-16-treated cells as compared to the control cells? We will observe the Hoescht fluorescence using a microscope filter set that is optimized for another DNA binding dye, DAPI, since Hoechst and DAPI have similar excitation and emission wavelengths. Your instructor will demonstrate how to use the fluorescence microscope during lab and how to take digital photographs of representative fields of the HL-60 cells. The files for these digital photos will be made available for you to use in your lab reports.
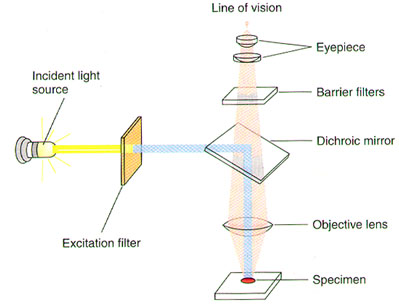
Figure 1. Optical System of a Fluorescence Microscope (Lodish H, et al. Molecular Cell Biology, p.140.)
Instructions for using the Fluorescent Microscope: Media:Using the Nikon 80i Fluorescence Microscope and Camera.doc
References:
Beltz, B.S. and G.D. Burd. (1989) Immunocytochemical Techniques: Principles and Practice. Blackwell Scientific Publications, Cambridge, MA.
Lodish H., Berk A., Zipursky S., Matsudaira P., Baltimore D. and Darnell J. (2000) Molecular Cell Biology, 4th ed. W.H. Freeman and Co., New York.
Watson, S.J. and H. Akil. (1981) Immunocytochemistry: Techniques, trials, and tribulations. Neurosci. Comm., 1: 10-15
Protocol
- 0.5x106 cells were pelleted and the supernatant discarded at each time point for control and etoposide (VP-16) treated cells.
- The cell pellets were mixed with 50 ul of 100 ug/ml Hoescht-33342 and incubated on ice for 5 minutes.
- The cells were then fixed with 10% formalin to preserve them for our use.
- The cells have been stored at 4°C
We will bring slides, coverslips, a P-20 pipette and tips to the microscope room. We will dispense 8 ul of fixed cells onto our side and cover it with a coverslip. We will then look at the cells to determine shape and size as well as degree of chromatin condensation. Hoescht dye emits at 492 nm and excites at 356 nm - similar to DAPI DNA staining.
Note: Not all cells will have condensed chromatin. It will be beneficial once we have taken a lot of pictures to go through them and determine the percentage of cells under each condition that have condensed chromosomes. I am expecting less than 50% for any time period.