BISC220/S13: Mod 1 Lab 3
Lab 1: Induction of β-galactosidase and Molecular Modeling
Lab 2: Affinity Chromatography and Specific Activity Determination
The Michaelis-Menten Model
Lab 4: Polyacrylamide Gel Electrophoresis
Media Recipes
Science Writing and Figure Design
Enzyme Kinetics
In this laboratory, the effect of substrate concentration on the reaction velocity of β-galactosidase will be measured in the presence and absence of an inhibitor. You will also determine the kinetic parameters Vmax, Km, and Ki for the reactions. To conduct these experiments you will use the partially purified β-galactosidase fraction obtained from the affinity chromatography protocol in Lab 2.
Enzyme Kinetics
Unlike most uncatalyzed reactions, the initial rate or velocity of enzyme-catalyzed reactions increases with increasing substrate concentration until a substrate level is reached beyond which further additions of substrate do not increase the initial rate. We measured specific activity of β-galactosidase last week under such saturating substrate conditions while varying the concentration of enzyme. This week we will use a single concentration of enzyme and measure velocity of the reaction using gradually increasing substrate concentrations. The maximal velocity (Vmax) will finally be achieved at substrate saturation of the enzyme. The substrate concentration that yields a velocity equal to one-half the maximum velocity is equivalent to the Km, or Michaelis-Menten constant, of the enzyme. The Km reflects the affinity of the enzyme for its substrate; a low Km value indicates high enzyme affinity for a particular substrate, while a high Km value indicates a lower affinity.
Vmax and Km may be influenced by the presence of other compounds in the assay mixture. In some cases the compounds act as inhibitors; in other circumstances, rate enhancement can be achieved. As part of today's experiment you will be determining the effect of a reported inhibitor, IPTG (isopropyl thio β-D-galactopyranoside), on the kinetics of β-galactosidase. You will see how analysis of the data obtained can be used to reveal if this compound is indeed acting as an inhibitor and if so, what type of inhibitor.
Protocol
Microsoft Word File: Media:Enzyme Kinetics Protocol rev 2010.doc
- You want to test an 80-fold range of ONPG substrate concentrations using a single concentration of purified β-galactosidase (PF). To decide what concentration of enzyme is appropriate, look back at your A420 measurements of the PF in the last lab. The dilution of the PF that gave an absorbance closest to 0.5 in 5 minutes of reaction time is close to the dilution you want to use, but not exactly. Remember that the PF you will use today has been diluted in glycerol, so the protein concentration would be lower if you made the same dilution. Calculate the new protein concentration in mg/ml of the PF in its present form and decide how it should be diluted to give you absorbance in the accurate range (0.1-1.0) over the entire range of substrate concentrations to be tested.
- Prepare 3.0 ml of the appropriate dilution of your purified fraction in Z buffer. Remember to mix the PF very well by vortexing before taking an aliquot for dilution. Label the enzyme dilution clearly and keep it and the PF stock solution on ice.
- Prepare 5 ml of a 1:10 dilution of the substrate ONPG. The stock solution is at a concentration of 4 mg/ml. Label the substrate dilution clearly. Keep the diluted and undiluted substrate on ice.
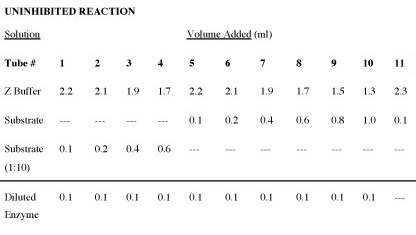
- To perform the assay without inhbitior, prepare 10 assay tubes and 2 reagent blanks (2 identical #11 tubes) in tubes using the volumes and solutions indicated in the table that follows entitled “Uninhibited”. DO NOT ADD ENZYME UNTIL YOU START THE REACTION.
- Equilibrate tubes 1-10 at 28°C for 5 minutes.
- Start the reaction by adding 100 µl (0.1 ml) of the diluted enzyme at 20 second intervals to each reaction tube but not the blanks (tubes #11). Vortex or invert to mix the reaction after the enzyme is added and return the tube to the water bath.
- Stop the reaction by adding 1.0 ml of 1 M Na2CO3 exactly 5 minutes after the addition of enzyme. Vortex each tube after adding stop buffer. Add stop buffer to the blanks at any time.
- Read your absorbances and make sure the enzyme dilution you used was appropriate before repeating the reactions with inhibitor. If the enzyme concentration is appropriate, absorbances will be largely within the linear range of the instrument and you will have clearly reached Vmax.
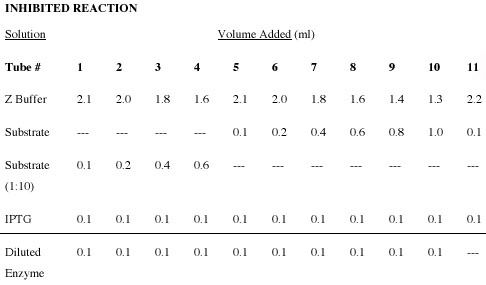
- To assay the effect of IPTG on the reaction, repeat the assay using a putative inhibitor of the enzyme, IPTG. Prepare another set of 11 test tubes using the volumes and solutions indicated in the table entitled “Inhibited” that follows. DO NOT ADD THE ENZYME AT THIS TIME.
- Equilibrate the 10 tubes at 28° for 5 minutes. Start the reaction by adding 100µl (0.1 ml) of enzyme dilution at 20 second intervals to each reaction tube (BUT NOT THE BLANKS). Stop the reactions exactly 5 minutes later by adding 1000µl (1.0 ml) of Na2CO3. Add stop buffer to the blanks too. Mix well by vortexing after each addition of enzyme or Na2CO3.
- Transfer all tubes to cuvettes making sure they are at least 2/3 to 3/4 filled. Zero the spectrophotometer at 420 nm using the reagent blanks. Read and record the absorbance of each assay.