BISC110/S13: Lab 7 Taster SNP2
Lab 7 - Taster SNP Week 2
Today you will finish determining your assigned DNA sample's genotype at the PTC gene locus by examining the SNP at nucleotide position 785. Last week you began the process by isolating bacterial plasmid DNA and setting up a PCR reaction to amplify a region of the PTC gene which contains the SNP of interest. Today you will digest your PCR product with the restriction enzyme Fnu4H1 and analyze the products by agarose gel electrophoresis.
PART I: Digestion of your PCR products with the restriction enzyme Fnu4H1
From the PCR reaction, you now have plenty of copies of the region of DNA we are interested in. To analyze the genotype of your sample, you will set up a restriction enzyme digest. How will this differentiate the SNP you have? Restriction enzymes are quite specific about the DNA sequences that they will cut. If the sequence is different, even by one base, the restriction enzyme will not cleave it. We have made use of this specificity in choosing a restriction enzyme, Fnu4H1, which cuts one allele of the PTC gene SNP, but not the other. Because the restriction digest will take an hour, you will set it up at the beginning of the lab period.
- Locate your PCR tubes from last week, and set them out at your bench to thaw.
- Each student will set up a restriction enzyme digest of her own PCR product and one student of each pair will also digest the PCR product of the heterozygous (T785/C785) control.
- Get a microfuge tube and label it with your ID number (from the course spreadsheet) and D to indicate that this will be the digested DNA. If you are doing the control, label another microfuge tube with a C and a D.
- Set your P20 to 10 μL and carefully pipette 10 μL of the appropriate PCR product into the microfuge tube(s) labeled with D. Be sure to save the remaining PCR product! You will also run this undigested DNA on your gel. Add the label U, for undigested, to the PCR tubes that contain the rest of the PCR product (after you have removed the 10 μL aliquot for digestion).
- Add 10 μL of restriction enzyme “cocktail” to the D tube(s) only. The "cocktail" contains Fnu4H1 and an appropriate buffer. (In each 20 μL reaction volume there is 2 μL New England Biological Buffer #4 (http://www.neb.com), 2.5 units (in 0.5 μL) of Fnu4H1 (New England Biological product RO171), 7.5 μL purified water and the remainder is template DNA). Mix well by pipetting up and down.
- Incubate your digests in the 37C heat block for 60 minutes.
DO NOT THROW AWAY ANY OF THE PCR TUBES!
PART II: Agarose gel electrophoresis.
View the animation of how to pour and run an agarose gel.
You can do several of these steps while you are waiting on your restriction digests.
We will be using a Biorad gel system. Each pair of students will use one of these systems.
Setting up the gel apparatus
- Place the gel-casting tray in the gel box on the raised platform, positioning the comb slots nearest to the negative electrode (Black).
- Place the metal wedges into the gel box in the slots on either side of the gel-casting tray.
- Place the comb into the slots closest to the edge of the gel-casting tray.
- Ask your instructor to check your set up.
Dilute the buffer and make an agarose solution
Each pair of students will make a 30 ml solution of 1.5% (w/v) agarose in 1X TBE (89 mM Tris Base (TRIZMA), 89 mM boric acid, 2 mM EDTA (pH 8.0).
In order to do this:
- You will be provided with a 10X TBE stock solution. You will need a total of 300ml of buffer for preparing your agarose solution and for running your gel. Calculate the amount of 10X TBE you will need in order to prepare 300ml of 1X TBE.
- Measure out _____ ml of 10X TBE
- Add_____ml of water.
- Use a stir bar to mix this 1X TBE solution (briefly) in a 500ml flask.
- Calculate the amount of agarose you should weigh out to make your gel.
- For 30 ml of a 1.5% agarose solution weigh out _____ g of agarose and put that in ______ ml of the 1X TBE buffer that you prepared.
- Check your calculations with your lab instructor before proceeding.
- Using one of the balances in lab, carefully weigh out the appropriate amount of agarose using one of the top loading balances and a piece of weighing paper. Transfer the agarose to the small, empty 125 ml Erlenmeyer Flask at your bench and add the proper amount of 1X TBE buffer. Swirl to mix. The agarose will NOT go into solution without high heat (the next step).
- Cover the open top of your 125 ml flask with the little beaker you found on it originally and microwave the agarose solution for ~15-20 seconds on high power. Use the “hot hands” to handle the flask when removing it from the microwave, since the glass will get very hot. The agarose should be completely melted and the solution should be uniformly clear when you swirl it around. If it isn't, microwave it again for a few more seconds (~10).
- Immediately take your flask to the instructor's bench and ask your lab instructor to add 3 μL of SYBR Safe stain to your flask. PUT ON GLOVES. Any time you work with your agarose or gel for the remainder of the lab, you will need to be wearing gloves.
- Place the flask on your bench and swirl the flask in a figure 8 pattern to gently mix for about 10-15 seconds. Then immediately proceed to the next step since everything is highly time sensitive at this point.
Cast a gel
If you pour the agarose while it is too hot, you can crack or warp the casting tray, which may cause leaking, but if the solution is too cool it will be a lumpy mess when you pour it and you will have to start all over again.
- When the lip of the glass flask is not too hot to hold with a bare hand (approximately one minute after you add the SYBR Safe), you may begin to pour the agarose into the gel-casting tray.
- Start out very SLOWLY pouring agarose into the CENTER of gel-casting tray, trying to avoid the formation of air bubbles. If you start in the center, by the time the agarose gets to the edges, it will be just cool enough to seal them without leaking. If you start out pouring VERY slowly, the casting tray is less likely to crack from the heat because the temperature can equilibrate incrementally. If bubbles form, remove them quickly before the gel starts to solidify by having your partner pop them with a Pasteur pipette.
- When you have poured the whole flask of molten agarose, allow it to set completely. When it has solidified, it will appear relatively opaque. Ask your instructor to check before you go on to the next step.
- Carefully remove the metal wedges from the gel box.
- Fill both sides of the gel box with 1X TBE running buffer, until the buffer is about 3 mm above the gel surface.
- To remove the comb from the gel, pull it slowly and straight up so that the wells are not damaged.
While you are waiting for your restriction digests to finish
Each student will run their own undigested and digested PCR product. You only have to run the digested control.
- Label a microfuge tube with your ID number and indicate that it contains undigested DNA (U).
- Pipette 10 μL of the appropriate PCR product from the PCR tube into the labeled microfuge tube.
- If you do not have 10 μL left, add as much of your PCR product as you have.
- Add 10 μL of water to each labeled microfuge tube. Mix thoroughly by pipetting up and down.
- Mix the loading dye thoroughly by pipetting up and down. Be sure to do this before you add loading dye to any tube. Once mixed, add 2.2 μL of loading dye to the microfuge tube. Loading dye contains the following components: 0.4 % Ficoll 400, 1.8 mM EDTA, 0.55 mM Tris-HCl, 0.00117 % SDS, 0.025 % Bromophenol Blue pH 8.0 @ 25°C)
- Don’t forget to use a clean, new tip for each tube.
- Your lab group should acquire a sample of DNA Ladder (ready to load) from your lab instructor. This contains DNA fragments of the following sizes: 766 bp, 500 bp, 300 bp, 150 bp and 50 bp. Using what you know about the SNP region we amplified with PCR and the location of the Fnu4H1 cutting site, you should be able to predict where you might see DNA fragments (looking like glowing bands) in relation to the position of the marker lane bands.
- Practice gel loading. Loading an agarose gel can be tricky. Your lab instructor will demonstrate how to use the practice dishes to learn to load a gel properly. Practice until you are comfortable and proficient.
Bromophenol blue: is a negatively charged dye that moves towards the positive pole.
Ficoll: is added to increase the density of the sample. This causes the sample to sink into the well, since it is heavier than the buffer.
EDTA: helps keep the nucleic acid in the sample stable by inhibiting nucleases that might be present. It does this by binding divalent cations, such as Mg2+, that are necessary for nuclease activity.
Do not throw the stock tube of loading dye away when you clean up after this lab!
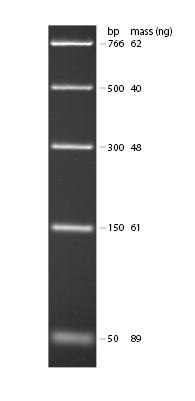
Figure 1. Example of DNA Ladder on 1.8% TBE agarose gel stained with ethidium bromide. Ladder from New England Biological (NEB3234).
Prepare your restriction digest for loading:
- After your restriction enzyme digest is complete, microcentrifuge your tubes briefly (5-10 seconds) to get any condensation from the lid into the bottom of the tube.
- Add 2.2 μL of loading dye to each tube of digested DNA (one tube for each member of your lab group, plus the heterozygous digested control).
- Each pair of students should now have 6 tubes which contain DNA and loading dye: one digested tube for each group member, one undigested tube for each group member, one digested control, and one tube of marker lane DNA.
Loading and Running the Gel
- Fill out the gel template provided so you know which lane each sample will be loaded into. Copy this template into your lab notebook. You will load 20 μL from each tube (including the DNA ladder) into separate gel lanes.
- Check your tubes before loading. If not all of the liquid is at the bottom of your tube, microcentrifuge briefly (around 15 seconds). Be careful not to let any bubbles into the pipette tip when you are drawing the sample into the tip. Give the template to your lab instructor.
- When all of the samples have been loaded, check the orientation of the gel with respect to the electrical leads. Remember “RED AHEAD”; DNA, having a net negative charge, will migrate toward the positive (red) electrode.
- Placing the lid on the box connects the leads to the gel box. Connect the other ends of the leads to the power supply. Check that the power supply is set at 100 volts, and then turn on the power. Observe bubbles rising from the electrodes at each end of the tank (more from the negative electrode than the positive electrode). After 1–2 min, observe the dye leaving the wells. Run the gels for approximately 30-35 minutes or until the tracking dye has moved about half way down the gel. Ask your instructor to check that your dye front has migrated sufficiently before you stop the electrophoresis by turning off the current.
Gel Imaging
- When the gel has separated your DNA fractions, disconnect the leads from the power supply and remove the lid from the gel rig.
- Wearing gloves, carefully remove your casting tray (still containing the gel) from the gel rig and transfer it to a labeled plastic sandwich box at your bench.
- Your instructor will take you to the instrument room next door and image your gel. The directions for the BioRad imager can be found in Appendix G.
- SYBR Safe stain is a fluorescent dye that binds to nucleic acids. It has excitation maxima at 280 nm and 502 nm, and an emission maximum at 530 nm. You will place your gel on a UV transilluminator, a device that shines ultraviolet light through the gel. Where nucleic acids have been stained with the SYBR Safe stain, a fluorescent green glow will be detected by the digital camera connected to the transilluminator.
- Your instructor will upload an image of your gel to your lab Sakai conference.
Analyzing the Gel
- Ascertain whether your enzymes cut your DNA. How can you tell? How many fragments do you see for each person in your lab group?
- Use the DNA ladder to estimate the size of the fragments you see.
Recording Your Data
- Determine your assigned genotype: C785/C785, C785/T785, or T785/T785. Be sure to check your interpretation of the gel with your lab instructor.
- Once you are sure of your assigned genotype, record your data in the class excel spread sheet. DO NOT LEAVE LAB UNTIL YOU HAVE RECORDED YOUR GENOTYPE ON THE TASTER EXCEL DOCUMENT!
Part III. Data Analysis & Science Writing Discussion
While your restriction enzyme digestion and electrophoresis are running, we will have time to talk about your expected results and their significance. We will also address how you might write about these data in the form of a primary research report due at the beginning of lab 9. This will be a partial paper (title, introduction, results, and references).
Assignment
1. We will use part of lab 8 as a data and writing workshop for the partial paper that will be due in lab 9. In preparation for the lab 8 workshop and discussion, read the Prince 2012 article. This article will be available as stated by your lab instructor.
2. Take the lab 9 pre-lab quiz.
3. After the last lab of the week, your lab instructor will post an edited version of the Excel document with genotype and phenotype data. Incomplete data will be removed from the datasheet since they do not add to the analysis. The following assignment (5 pts) should be done individually:
a. Please generate at least two figures to address the following question: Is there a correlation between age of P. aerugonisa acquisition in cystic fibrosis patients and the TAS2R38 genotype data?
b. Make an outline (with bullet points) of what you plan to include in your introduction and results sections.
c. Upload an electronic version of your figures and outline to Sakai. Your instructor will give you more specific details about where to upload the files. In addition, please bring a hard copy of your figures and outline to lab 8.
In the partial paper due in lab 9, you will be required to cite at least 6 papers in your introduction.