BISC110/S13: Lab 6 Taster SNP1
Introduction
Single nucleotide polymorphisms (SNPs, pronounced “snips”) represent the simplest type of genetic variation between individuals. A SNP refers to a specific location in the genome where different people have been shown to have a different nucleotide. If more than one percent of the population has a different nucleotide at a particular location, that location is considered a SNP. If less than one percent of the population has a different nucleotide, it is considered a mutation (Alberts et al. 2010).
There are approximately 3 million SNPs throughout the human genome, yet they do not all behave independently. Groups of nearby SNPs are often inherited together in what are called haplotype blocks (see Figure 19-36 in your Alberts text). Depending on the region of the genome and the location of prior meiotic recombination, a haplotype block can span one or many genes.
Some SNPs appear to be benign, while others are associated with an increased risk of developing disease. To try to determine genetic risk factors for disease, scientists have compared SNPs in healthy individuals to SNPs in individuals with a particular disease. If a SNP is associated with an increased risk of disease, it’s important to keep in mind that it may not be causative. Furthermore, many SNPs that are associated with disease confer only a modest increase in disease risk. In this lab, you will have the opportunity to experimentally examine a SNP located within the TAS2R38 gene (also called the PTC Taster gene). While variations in TAS2R38 genotype had long been thought to be benign, recent work has suggested a correlation between TAS2R38 genotype and disease risk (Carrai et al. 2011, Lee et al. 2012).
PTC Taste Genetics
In the 1930’s Arthur Fox, a chemist at Dupont, synthesized phenylthiocarbamide (PTC). While transferring the compound to a bottle he generated a cloud of PTC dust which elicited a dramatic response in his lab mate. Although Fox tasted nothing, his colleague, C. R. Noller, complained that the dust was intensely bitter. (If you are interested, Arthur L. Fox’s original 1932 paper, The Relationship Between Chemical Constitution and Health, can be read at the web site of the journal, The Proceedings of the National Academy of Science.)
Since Fox's serendipitous discovery, variation in the ability to taste PTC (and other related bitter compounds such as those found in broccoli!) has become one of the most studied human genetic traits. On the surface, PTC sensitivity appears to be inherited as a simple Mendelian trait with two alleles, T for taster and t for nontaster. In reality its inheritance is much more complicated. Today we know that PTC sensitivity is mediated by the PTC Taster gene (aka TAS2R38) that encodes a bitter taste receptor (a heteromeric G-protein coupled receptor) which is found on the surface of cells of the tongue (Kim et al. 2003).
Additional studies of the PTC Taster gene in humans across the globe have revealed that there are two common alleles, taster and nontaster; however, there are at least nineteen other rare alleles that affect the taster phenotype. These different forms of the gene code for proteins that vary in their ability to bind bitter compounds such as PTC. The two common alleles of the PTC Taster gene differ by three SNPs (see table below).
| Nucleotide position | Taster | Nontaster |
|---|---|---|
| 145 | CCA pro |
GCA ala |
| 785 | GCT ala |
GTT val |
| 886 | GTC val |
ATC ile |
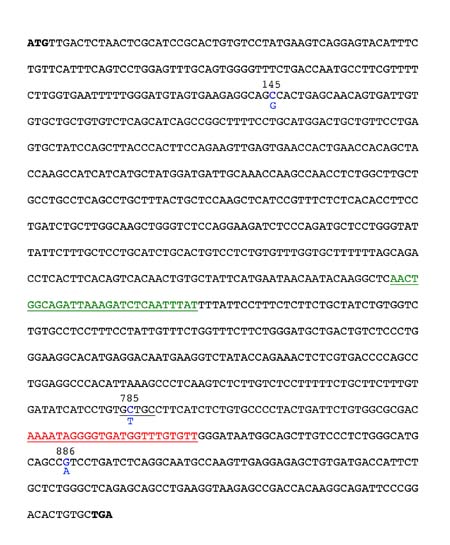
The common nontaster allele has a G at nucleotide position 145 (G145), T at position 785 (T785) and A at position 886 (A886). This nontaster allele (G145/T785/A886) produces a polypeptide with alanine, valine and isoleucine at these sites and therefore is referred to as the AVI allele. The common taster allele C145/C785/G886, produces a polypeptide with proline, alanine, and valine at these sites and is referred to as the PAV allele.
The SNP at position 785 is useful for determining PTC genotype since the common taster sequence in this region forms a Fnu4H1 restriction enzyme digestion (cut) site. In contrast, in the common nontaster allele, there is a T at position 785 (instead of C785), which eliminates this restriction enzyme digestion (cut) site. A restriction site is a specific DNA sequence that is recognized and cleaved by a specific restriction enzyme or endonuclease. There are two other Fnu4H1 restriction sites within the PTC gene, but we will use primers specifically designed to amplify the region flanking the one polymorphic Fnu4H1 site at position 785 while excluding the other two. Using these primers in a polymerase chain reaction (PCR) generates a 303 base pair (bp) DNA fragment. Restriction digestion of this PCR product with Fnu4H1 will yield one 303 bp fragment for the common nontaster allele (because the restriction enzyme cut site is not present) and two shorter fragments (64 bp and 239 bp) for the common taster allele. Visit the Dolan DNA center web site to learn more about PCR and see an animation of how it works.
Fnu4H1 Restriction Site
5’-GCNGC-3’
3’-CGNCG-5”
N can be any of the 4 bases
Figure 1. Sequence of the common Taster allele of the PTC gene. The 3 SNPs are highlighted in blue with the nucleotide found in the common nontaster allele shown below the taster. The position of the forward and reverse PCR primers are shown in green and red, respectively. The Fnu4H1 site that will be present in your PCR products and which you will exploit for genotype analysis is underlined in black surrounding position 785.
In two-weeks of lab experiments and a third lab meeting of discussion, you will be provided with a phenotype and determine the genotype at the PTC Taster gene locus of a hypothetical cystic fibrosis patient. You will also explore some of the ethical issues surrounding personal genomics and learn more about writing a scientific paper. Since acquisition of P. aeruginosa is associated with health problems in cystic fibrosis patients, you will be assigned a phenotype and determine the PTC genotype of this hypothetical patient. This will allow you to determine if there is any relationship between the PTC genotype and age of P. aerugonisa acquisition. In the first week you will be given a sample of human DNA containing a section of the PTC gene. You will begin the process of determining your sample’s genotype by extracting and purifying human DNA contained within bacterial plasmids (a plasmid is a circular piece of DNA). You will then use the polymerase chain reaction (PCR) to amplify a region of DNA that contains a SNP in the PTC Taster gene. PCR is a technique which exploits in vitro DNA replication by a heat stable DNA polymerase to produce millions of copies of a specific DNA sequence. In addition, we will watch a video about some of the ethical issues related to the use of SNPs and other genetic information. In the second lab you will experimentally determine your sample’s genotype by digesting your PCR product with the restriction enzyme Fnu4H1 and analyzing the products by agarose gel electrophoresis. You will then analyze phenotype and genotype data from the entire course and make figures in preparation for a data analysis meeting.
Lab 6 - Taster SNP Week 1
PART I: DNA extraction and purification (using QIAprep® Spin Miniprep Kit from Qiagen)
You will use a kit to isolate bacterial plasmid DNA (containing part of the human PTC gene). You will use a commercially available kit to isolate bacterial plasmid DNA (containing part of the human PTC Taster gene). You will chemically break these bacterial cells open and use a silica-based spin column to separate the DNA from the other cellular components.
- Your instructor will assign you a DNA sample number and tell you the age of acquisition of P. aeruginosa. Record your DNA assigned number and phenotype in your lab notebook, so you don't forget them! You will need to remember your number so you can record your genotype results next to the phenotype results you have been given.
- You will be provided with a bacterial cell pellet in a 1.5 mL microfuge tube. Be sure that this tube has the same number that you were just assigned.
- Resuspend the pelleted bacterial cells in 250 μL of Buffer P1 (resuspension buffer) by vortexing or pipetting up and down until no cell clumps remain. Check with your instructor to ensure that you have resuspended the cells properly.
- Add 250 μL of Buffer P2 (lysis buffer) and mix thoroughly by gently inverting the tube 6 times. Do NOT vortex or the DNA will be sheared. Be sure that the solution is viscous and slightly clear before proceeding to the next step. In any case, do not let the lysis reaction proceed for more than five minutes.
- Add 350 μL of Buffer N3 (neutralization buffer) and mix immediately and thoroughly by inverting the tube 6 times. The solution should become cloudy.
- Centrifuge for 10 minutes at 13,000 rpm in a table-top microcentrifuge. A compact white pellet will form along the side of the tube.
- Label a QIAprep spin column with your number while you are waiting for the centrifugation step to be done.
- Apply the supernatant from the step 6 to your labeled QIAprep spin column by decanting.
- Centrifuge for 30 seconds at 13,000 rpm and discard the flow-through by dumping the contents of the collection tube into your liquid waste container.
- Wash the QIAprep spin column by adding 500 μL of Buffer PB (to wash away trace nucleases) and centrifuging at 13,000 rpm for 30 seconds. Discard the flow-through by dumping the contents of the collection tube into your liquid waste container.
- Wash the QIAprep spin column by adding 750 μL of Buffer PE and centrifuging at 13,000 rpm for 30 seconds. Discard the flow-through by dumping the contents of the collection tube into your liquid waste container.
- Centrifuge the QIAprep spin column for an additional 1 minute to remove any residual wash buffer. Residual ethanol from Buffer PE may inhibit the PCR reaction so it is very important that you discard the flow-through in step 11 before proceeding with this step.
- Discard the collection tube and place the QIAprep column in a clean labeled 1.5 ml microcentrifuge tube. To elute the DNA from the QIAprep column, add 50 μL Buffer EB (10mM Tris-Cl, pH 8.5) to the center of the QIAprep spin column. Be careful not to touch the membrane with your pipette tip. Let stand 1 minute and then centrifuge at 13,000 rpm for 1 minute. You now have purified bacterial plasmid DNA containing a region of the human PTC Taster gene.
PART II: Dilution of your DNA for use in the PCR reaction
You probably purified between 5 and 15 μg of plasmid DNA! Since it is ideal to use less than 1 ng of plasmid DNA in a PCR reaction, you will need to make a 1/10,000 dilution of your DNA in water. You could add 1 μL of plasmid DNA to 9,999 μL of water, but this would not be a very accurate way to make a 1/10,000 dilution. Therefore, you will set up a series of dilutions to obtain your desired final dilution.
- Begin by making 1000 μL of a 1/100 dilution of your plasmid in water. To do this, you should add _____ μL of plasmid DNA to ______ μL of water. Using your 1/100 dilution, then make 1000 μL of a 1/10,000 dilution by adding ______ μL of your 1/100 dilution to _______ μL of water. You now have a 1/10,000 dilution of plasmid DNA and you will use this in PART III.
- Store your DNA on ice until you are ready to set up the PCR reaction.
PART III: PCR of your PTC Taster gene (using PuRe Taq Ready-To-Go Master Mix beads (GE Healthcare)
Next, you will set up a polymerase chain reaction (PCR) designed to amplify a portion of your extracted DNA. Specifically, we will amplify the region of the PTC Taster gene that contains the SNP at position 785 (the one which creates a Fnu4H1 site if a C is present). We need to use PCR so we will have enough DNA to specifically analyze the region of the PTC Taster gene surrounding the SNP at position 785. The forward and reverse primer sites (where the primers will bind) for our reactions are shown in Figure 1. In your notebook record the sequences of the forward and reverse primers. Use the convention of writing the primer sequence beginning with the 5’ end. As you determine the primer sequences, keep in mind that your template DNA will initially be double stranded (just one of the strands is shown in Figure 1). The strand shown in Figure 1 is represented in the 5' to 3' orientation. Imagine that the complementary strand is also there (the complementary strand must be in the 3' to 5' orientation since the two DNA strands are antiparallel). The two template DNA strands will separate during the denaturation phase of the PCR reaction and then the primers will be able to anneal during the annealing phase of the PCR reaction. To determine the orientation of the primers, keep in mind that newly synthesized DNA will be made in the 5' to 3' direction (therefore new nucleotides will be added to the 3' end of the primer) during the extension phase of the PCR reaction. Have your instructor check your sequences before proceeding.
- Locate a PCR tube, containing a master mix bead. Use a permanent marker to label the top of the tube with your student number. One partner in each group will also prepare a control DNA sample for PCR in addition to her own DNA. She will obtain 2 PCR tubes and label one with her student number and the other will be labeled C (for Control). The control DNA contains a mixture of the C785 and T785 alleles. Marvel at the size of these tubes. “Master mix” is the commonly used term for this subset of the PCR components: it contains Taq polymerase, dNTPs, and buffer. In this case, the components have been dried into a bead, so you’ll dissolve the bead into solution using your primers and DNA. Be especially careful with the PCR tubes. The walls are very thin and easy to damage.
- Tap the bottom of the tubes, very gently, on your bench to get the bead of reagents into the bottom of the tube BEFORE you open it! Pipette 20 μL of primers into each PCR tube. The two primers (one forward and one reverse primer) necessary to replicate both strands of your DNA are included in this reagent. The final concentration of each primer in the PCR tube will be 0.2 μM.
- Very carefully, pipette 5 μL of your 1/10,000 dilution of plasmid DNA into the appropriate PCR tube. If you are doing the control, go to the instructor's bench and ask him/her to pipette 5 μL of control DNA into your PCR tube labeled C.
- Mix the contents well by gently flicking the bottom of the tube. Then, vortex the tubes gently (at a vortex speed of 3) until the beads have gone completely into solution. The contents of the tubes should be clear.
- Using the centrifuge located next to the PCR machine, microfuge the tubes briefly (5 seconds) to get all the contents to the bottom. You will need to use adapters to microfuge these tiny tubes; the microcentrifuge next to the PCR machine is outfitted with these adaptors.
- Follow the instructions of your lab instructor for loading your samples into the PCR machine (called a “thermal cycler”). Your instructor will tell you how to record the location of your tubes in the thermal cycler on the template provided.
- Your lab instructor will return to lab after the PCR cycles are complete and transfer your PCR reactions into the freezer for storage.
Here is the cycle we will use for your samples:
First, there is an initial step for 10 minutes at 95C to activate the polymerase.
This is followed by 40 cycles with the following pattern:
1 minute at 95C, for “melting” the DNA
1 minute at 55C, for annealing the primers to the single stranded DNA
1 minute at 72C, for synthesizing new DNA
There is then a 10 minute stage at 72C, to complete whatever synthesis has been started.
Finally, the tubes are chilled at 4C until someone moves them to the freezer for longer term storage.
PART IV. SNPs and bioethics
- As a class we will view a short video of a Nova scienceNOW episode that explores some of the ethical and policy issues that stem from the use of SNPs and other forms of genetic testing for medical diagnosis and assessment of genetic risk. Movie can be accessed at : NOVA Science NOW
Assignments
- Read the following background information on restriction enzymes found at: SNP Restriction Enzymes and on gel electrophoresis at: Gel Electrophoresis
- Read the Hall 2010 article.
- Take the Lab 7 pre-lab quiz.
- (10 points) Answer the following SNP Taster Questions. This assignment must be done individually.
The Answers to the following questions will be turned in though Sakai before the start of Lab 7.
1. Ingrid has the following SNPs on one copy of chromosome 20:
G at position 334 of gene A
T at position 586 of gene A
C at position 23 of gene B
A at position 437 of gene B
A at position 978 of gene B
Determine if the following combinations of SNPs are considered to be a haplotype only, an allele only, or both.
______________________________ G334, T586
______________________________ C23, A437, A978
______________________________ G334, T586, C23, A437, A978
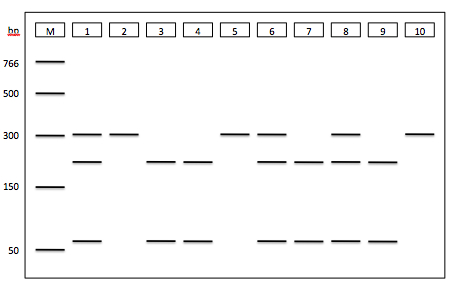
2. A section of the TAS2R38 gene (spanning the SNP at position 785) was amplified by PCR using DNA from 10 individuals as templates. The PCR products were digested with Fnu4H1 and the digested PCR products were separated by agarose gel electrophoresis. Use the results from the gel below to answer the following questions (2 and 3).
Highlight the correct genotype for each individual.
Individual #1: C785/C785 C785/T785 T785/T785
Individual #2: C785/C785 C785/T785 T785/T785
Individual #3: C785/C785 C785/T785 T785/T785
3. You are interested in investigating whether the SNP at position 785 of the TAS2R38 gene is associated with susceptibility to sinus infections. Use the gel electrophoresis data (from question 2) and the phenotype data (below) to determine if there is an association between the 785 SNP and sinus infection.

You should use the gel electrophoresis and phenotype data to generate a figure that will allow you to address this question.
4. Based on the figure, is the SNP at position 785 associated with susceptibility to sinus infections? Your response should be one paragraph.