BIOL368/F14:Isabel Gonzaga Week 8
Defining Your HIV Structure Research Project
This research project will be completed in conjunction with Nicole Anguiano and Chloe Jones.
Question
How does HIV status (diagnosed, progressing or non-trending) affect the structure of the V3 protein region?
Hypothesis
We hypothesize that diagnosed groups will express greater variability in the V3 region in their protein structure, in comparison to the non-trending groups. Initial comparisons show that diagnosed groups and progressing groups expressed greater genetic variability than non-trending groups. These changes may affect the third variable region, affecting the host's ability to adapt to the changes and generate sufficient immune response.
Subject Data
According to the BEDROCK HIV Sequence Data Table, I was able to determine which of the subjects used within my study actually developed aids. All 3 AIDS diagnosed were confirmed with the disease by their final visit. In the AIDS progressing groups, subjects developed AIDS within 1 year after their final visit. The Non-Trending groups all maintained high CD4 T Cell Counts above the threshold, even after the study was conducted. Sequences were for each visit and subject were chosen using a Random Integer Generator, to eliminate selection bias.
The following sequences was taken from the BEDROCK HIV Problem Space Database, from the Markham et al. (1998) study.
Table 1: Sequences analyzed
| Group | Subject | Visit | Sequences |
|---|---|---|---|
| AIDS Diagnosed | 3 10 15 |
1 6 1 6 1 4 |
1, 2, 4 3, 4, 5 3, 6, 7 2, 4, 8 2, 3, 4 5, 8, 10 |
| AIDS Progressing | 7 8 14 |
1 5 1 7 1 9 |
2, 3, 9 2, 8, 9 1, 4, 5 1, 6, 7 2, 3, 4 9, 10, 11 |
| No Trend | 5 6 13 |
1 5 1 9 1 5 |
1, 3, 12 4, 8, 9 3, 4, 10 6, 7, 9 1, 3, 8 3, 5, 7 |
Working with Protein Sequences In-class Activity
Source: Bioinformatics for Dummies pp. 110-123;
Analysis using the HIV gp120 envelope protein
Chapter 4: Reading a SWISS-Prot Entry
- The Uniprot (previously known as SWISS-Prot) database was accessed
- Searching HIV generates 600,415 results
- Searching gp120 generates 182,286 results
- Accession number "Q75760", which corresponds to the the HIV gp120 structure used for Huang et al. (2005), was searched for and accessed on the database
- General Information about the entry
- Entry name: Q75760_9HIV1
- Primary Accession Number: Q75760
- Integrated into UniProt: November 1, 1996
- Sequence last modified: November 1, 1996
- Annotations last Modified: October 1, 2014
- Name and Origin of the Protein
- Protein Name: Envelope glycoprotein gp160
- Gene Name: env
- From: Human immunodeficiency virus 1; Taxonomic identifier 11676
- Taxonomy: Viruses › Retro-transcribing viruses › Retroviridae › Orthoretrovirinae › Lentivirus › Primate lentivirus group
- Publications
- 9 listed
- The Cross-References
- EMBL: Coding Sequence U63632.1
- PDB Crystal Structures: Entry 2b4c
- Modbase: theoretically calculated model database
- DIP (Database of Interacting Proteins): DIP-59960N
- IntAct: Q75760. 3 interactions observed
- Interpro
- PFam
- The Keywords
- Apoptosis
- Fusion of virus membrane with host membrane
- Host-virus interaction
- Viral attachment to host cell
- Viral immunoevasion
- Viral penetration into host cytoplasm
- Virus entry into host cell
- The Features
- Glycosylation at residue 298 (1 aa long); n linked
- 'Feature viewer' not active
- The Sequence
- Q75760 was downloaded in FASTA format
- General Information about the entry
Chapter 5: ORFing your DNA sequence
Taken from Bioinformatics for Dummies, 2ed. pp 146-147
- NCBI Open Reading Frame Finder was accessed
- HIV gp120 envelope protein sequence data for Subject 9 Visit 1 Clone 2 copied and pasted into the ORF input box. 'OrfFind button' was clicked.
- This translates the DNA sequence to the amino acid sequence and helps to locate the open reading frame (likely +1)
- ExPasy Translate was used to further analyze presence of open reading frame. Likely determined to be 5'3' Frame 1.

Chapter 6: Working with a single protein sequence
Taken from Bioinformatics for Dummies, 2ed. pp. 159-195. Analyzed using HIV gp120 env protein, sequence from UniPort.
Predicting the main physico-chemical properties of a protein
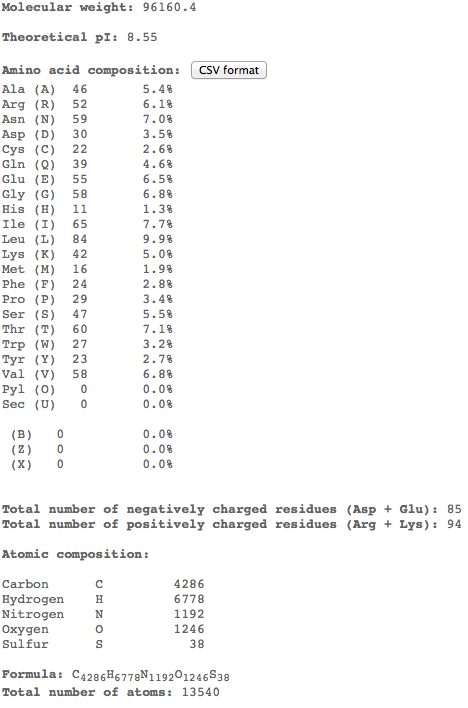
- ExPasy ProtParam was accessed
- Accession number Q75760, which corresponds to the the HIV gp120 structure used for Huang et al. (2005), was inputted
- Molecular Weight: 96160.4 Da (theoretical)
- Extinction Coefficients:
- Assuming all Cys form cystines: 184145 M1cm1
- Assuming all Cys mols are reduced: 182770 M1cm1
- Stability Index: 37.91 (stable)
- Half-Life:
- 30 hours (mammalian reticulocytes, in vitro)
- >20 hours (yeast, in vivo)
- >10 hours (Escherichia coli, in vivo).
Digesting a protein in a computer
- ExPasy Peptide Cutter tool was accessed and Q75760 accession number was inputted
- The following enzymes were selected (arbitrarily) for cleavage: Arg-C proteinase, Pepsin (pH1.3), Proteinase K)
Doing primary structure analysis
Looking for transmembrane segments
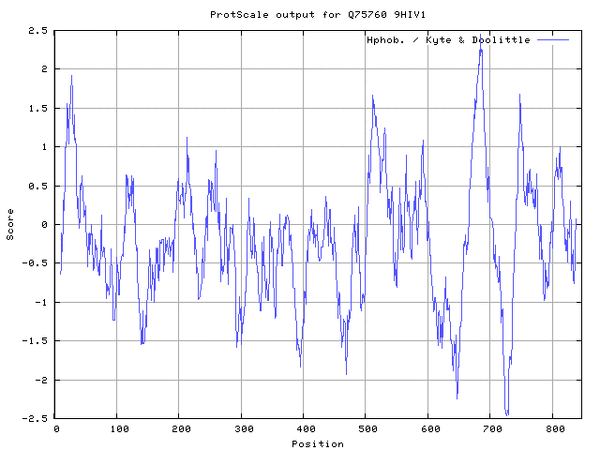
- ExPasy Protscale was accessed and Q75760 accession number was inputted
- Hphob. / Kyte & Doolittle selected, window size set to 19
- Results were converted to .gif format
- 4 transmembrane domains identified
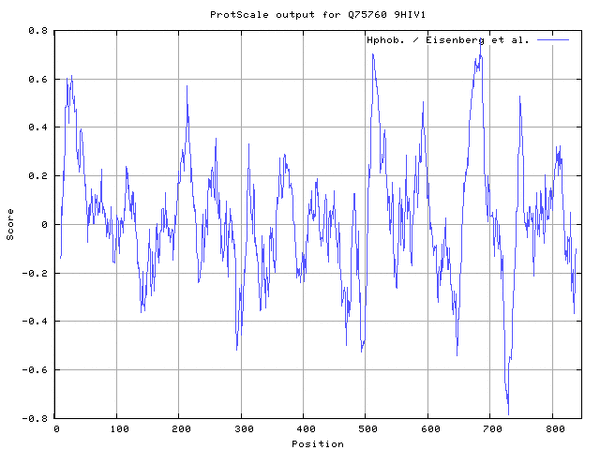
- Repeated Protscale analysis using Eisenberg et al. to confirm findings
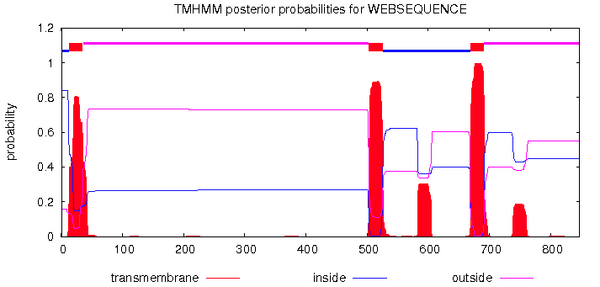
- TMHMM page of CBS site accessed, and Q75760 sequence was inputted in FASTA format
- 5 transmembrane domains observed
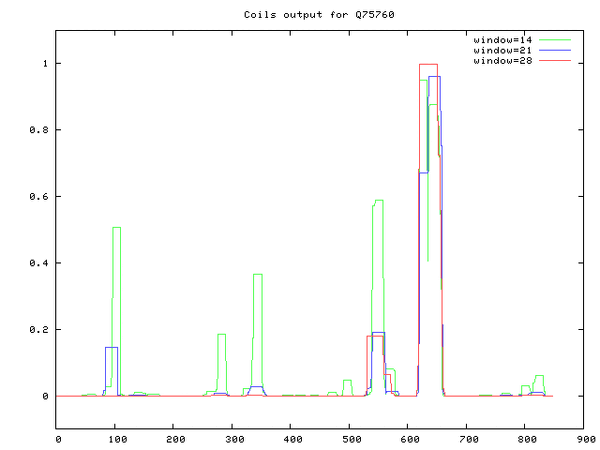
Looking for coiled-coil regions
- COILS server at EMBnet accessed and Q75760 sequence ID inputted
- Coil-coil regions determined at ~5 places (largest between 600-700 aa residues)
Predicting post-translational modifications in your protein
- ScanProsite accessed and Q75760 Accession Number inputted
- Unchecked exclude motifs with a high probability of occurrence box; checked do not scan profiles
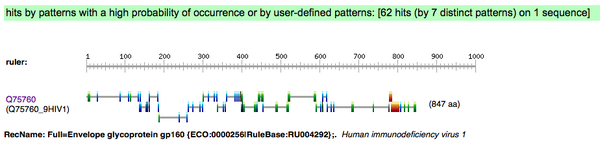

Finding Known Domains in your Protein
InterProScan
InterProScan was accessed and the FASTA sequence for glycoprotein gp160 was entered in the text box. Run was submitted.

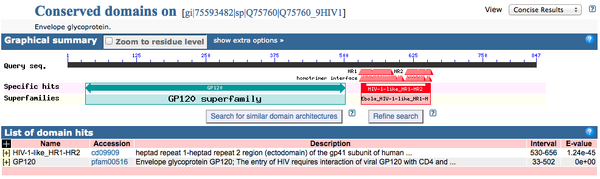
CD-Search
NCBI CD Server was accessed and Q75760 accession number was inputted. Expect Value Threshold set to 1 and 'Submit Query' button clicked.
Motif-Scan
Motif-Scan was not accessible via the provided website
Chapter 11: Working with Protein 3D Structures
Taken from Bioinformatics for Dummies, 2ed. pp. 330-336.
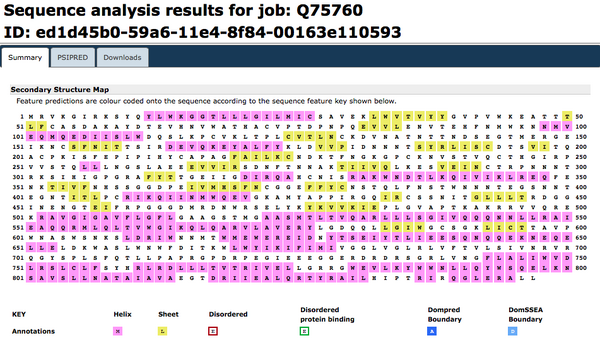
Predicting the secondary structure of a protein sequence: PsiPred
PsiPred was accessed. FASTA amino acid sequence for the Q75760 accession number was inputted.
Predicting additional structural features: PredictProtein
PredictProtein was accessed. A personal account was created and validated. The FASTA amino acid sequence for Q75760 accession number was inputted.
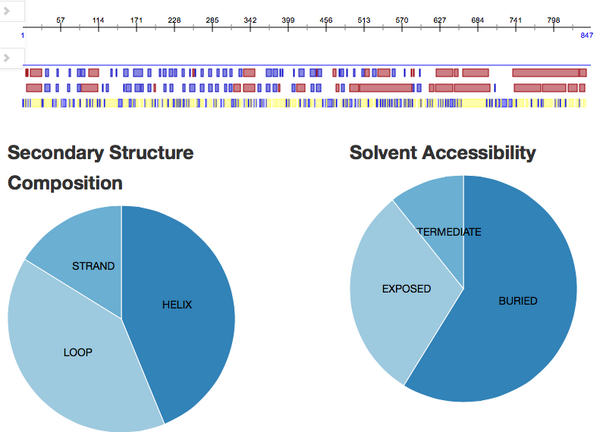
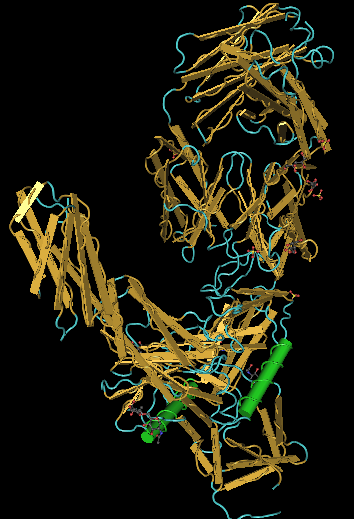
Crystal Structure Comparison
CN3D file was downloaded for the Hunag et al. (2005) crystallized protein from the NCBI website. Alpha helices were colored in green and beta sheets were colored yellow, using the coloring shortcuts. The true structure of the protein shows it to be predominantly composed of beta sheets of random coil, despite the prediction that alpha helices would be more prevalent (according to PsiPred).
Weekly Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 13 Assignment
- Week 15 Assignment
Class Journals
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 13
- Class Journal Week 15