BE.109:Systems engineering/Device characterization
Introduction
In an ideal world, each genetic part, device and system would come with a specification sheet. Since experimental variations can affect a part’s performance, specification sheets would include information about the part’s “operating conditions,” e.g. the optimal strain, media, temperature, and growth state, or perhaps in a few years, the community will recognize a set of conditions as “standard.” It would be wonderful if, knowing these conditions and the parts specifications, a user could predict the behavior of a genetic element, anticipating the amount of DNA, RNA and protein generated rather than taking the current approach, namely to try the experiment and find out. There are many methods to quantify DNA copy number, RNA levels and a protein’s concentration, activity, and interactions, so it would be ideal if the specification sheet reported values that held true no matter which measurement technique was used. And while we’re dreaming, why not also include information to help combine genetic elements in sensible ways, detailing a part’s incompatibilities or the level of crosstalk between parts at a minimum.
The challenge in characterization is to decide what aspects of a biological response need to be known and then to decide how best to measure them. Different measurement techniques are sensitive to different things. Since no description of engineering efforts would be complete without a car analogy, consider the problem of “noise” in biological responses in terms of what you’d see at a traffic light on a busy road that’s 10 lanes wide. At the red light the cars will wait, all poised to go forward. As the light turns green, one of the cars may race ahead, another may move ahead slowly. If a driver is distracted with the radio or a cell phone call (or lost in thought about the BE.109 “for next time” assignment), that car may not move until a nearby car moves or honks. Thus, even if all The cars get the same green light signal at the same time, they don’t perform identically. So what can be predicted about the reaction of cars (and their drivers) to a green light? As a population they move forward, individually they do so with different rates and if you were to look at just one car over time you might observe a different response than if you looked at another.
Biological responses are similarly noisy and inherent fluctuations lead to the heterogeneity of a cell population. As an example, consider again the “repressilator” plasmid that was introduced earlier (Elowitz and Leibler, Nature, 2000). The design, which mimics an electronic oscillator, should generate changing concentrations of three proteins (cI, tetR, and lacI), and flash GFP as a reporter for the tetR concentration. Simulations showed distinct peaks of the three proteins, temporally oscillating as intended, when modeled with a half dozen, coupled first-order differential equations. Periodicity was seen even when the system was modeled with more complex terms to account for confounding factors like cooperative binding, leaky promoters and different rates of mRNA decay.
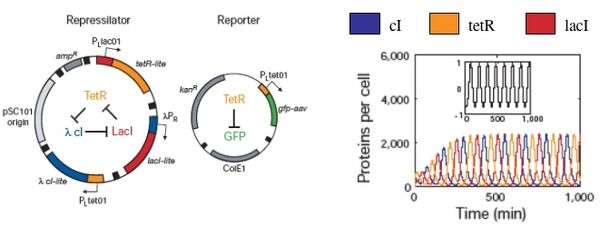
When the cells were built and tested, the GFP production fluctuated as the simulations predicted, but, perhaps surprisingly, not in every cell and not in unison. GFP oscillation was observed in ~ 40% of the cells (leaving ~60% of the cells not behaving as predicted), and for those that did flash, the period was measured as 160 minutes +/- 40. This observed period is longer than predicted by modeling, and showed a large standard deviation, with sibling cells exhibiting the most highly correlated behavior but only for a few cell divisions. So while this work provides an important demonstration of “cells built to order” it also reveals some inherent limitations.
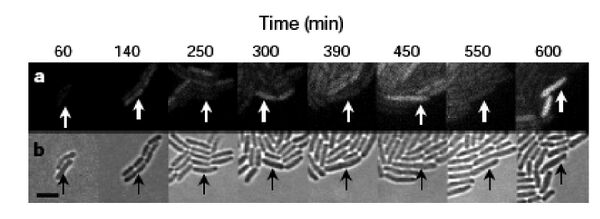
As you consider the measurements you are making in lab, it’s important to know the strengths and limits of the techniques you’ve chosen. Noise is most evident when observing the behavior of individual cells within a population, like the microscopic observations detailed above. However, single-cell measurements are generally time and labor intensive and there are limited ways to count individual molecules in a single cell. More common are population measurements, for example beta-galactosidase activity data, however these bulk measurements cannot differentiate hetero- from homogeneous expression. 10% of the cells expressing LacZ at 100% efficiency will give the same activity measurement as 100% of the cells expressing LacZ at 10% efficiency. Distributions within a population can be measured with techniques like FACS and flow cytometry, but it is difficult to apply these techniques to study individual cells over time.
Without a genie to grant us all our wishes, we are left with a somewhat limited menu of choices for system and parts characterization. Nevertheless, it’s worth considering what a perfect characterization would look like, informing this description with user experiences. Collectively this experience can help define some uniformly satisfying standards for parts and systems characterization. Today you will be generating some useful data on the robustness of the bacterial photography system, measuring its protein and RNA output under different operating conditions. You will be completing the experimental work on this characterization next time. The data analysis and documentation of your work will be done in the final lab session of this experimental module.
Protocol
Part 1: Measure cell number
Last time, you cultured the bacterial photography system under two experimental conditions. You should begin today’s lab by moving the bacterial culture to two 15 ml falcon tubes then spectrophotometrically measuring the number of cells in each population.
1. Make 1:10 dilution (100 μl into 900 μl Zbuffer –BME) of each culture.
2. Use 0.5 ml to measure the optical density at 600 nm for each dilution, using Zbuffer-BME to blank the spectrophotometer. Save the remaining dilution on ice to use in Part 2.
| OD600nm | Cells/ml (using 1 OD600~109 cells/ml) | |
|---|---|---|
| Standard growth condition | ||
| Variable growth condition |
Part 2: Beta-gal assay
Refer back to the protocol in the Protein Engineering Module to help you perform these assays.
| Sample | # | OD600nm | Time in (mins:secs) | Time out (mins:secs) | Elapsed time | OD420nm | OD550nm | Units |
|---|---|---|---|---|---|---|---|---|
| Standard | 1 | 0:00 | ||||||
| 2 | 0:10 | |||||||
| 3 | 0:20 | |||||||
| Variable | 4 | 0:30 | ||||||
| 5 | 0:40 | |||||||
| 6 | 0:50 |
Part 3: Isolate DNA
We will be using the number of cells as a reference point for our RNA measurements, assuming that each cell has one copy of its genome and, consequently, one copy of the LacZ reporter gene. From the data you collected in Part I of today’s experiment you already know the number of cells. Here you will lyse the cells to release that number of LacZ DNA pieces into the solution.
1. Move 1 ml of cells to eppendorf tube, spin, remove supernatant.
2. Wash cells by resuspending them with 1 ml PBS, resuspend in 1 ml PBS.Use 500 μl to read OD600nm.
3. Dilute to 10*8 cells/ml in PBS and add 1 μl of this dilution to a PCR tube with 9 μl lyse-n-go.
4. Run Lyse-N-Go program on PCR.
| Cycle # | Temp (°C) | Time (seconds) |
|---|---|---|
| 1 | 65 | 30 |
| 2 | 8 | 30 |
| 3 | 65 | 90 |
| 4 | 97 | 180 |
| 5 | 8 | 60 |
| 6 | 65 | 180 |
| 7 | 97 | 60 |
| 8 | 65 | 60 |
| 9 | 80 | hold |
3. Freeze your samples at –80°C until next time.
Part 4: Isolate RNA
RNA is strikingly different from DNA in its stability. Consequently it is more difficult to work with RNA in the lab. It is not the techniques themselves that are difficult; indeed, many of the manipulations will seem identical to those used for DNA. However, RNA is rapidly and easily degraded by RNases that exist everywhere. There are several rules for working with RNA. They will improve your chances of success. Please follow them all.
- Use warm water on a paper towel to wash lab equipment, like microfuges, before you begin your experiment. Then wipe them down with “RNase-away” solution.
- Wear gloves when you are touching anything that will touch your RNA.
- Change your gloves often.
- Before you begin your experiment clean your work area, removing all clutter. Wipe down the benchtop with warm water then “RNase-away,” and then lay down a fresh piece of benchpaper.
- Use RNA-dedicated solutions and if possible RNA-dedicated pipetmen.
- Start a new box of pipet tips and label their lid “RNA ONLY.”
- Move 1 x 109 cells from each cell culture to an RNase-free eppendorf tube.
- Spin the tubes for 1’ in a microfuge at full speed. Pour out the supernatant and flick the last drops of media out.
- Resupend the cells in 100 μl H2O with Lysozyme and incubate at room temperature for 5 minutes.
- Add 350 μl RLT with BME and mix by vortexing after each addition.
- Collect two Qiashredder columns from one of the teaching faculty. The lysates must be passed through these columns to remove particulate matter. Load the top of each Qiashredder column with the cell lysates. Microfuge the Qiashredder columns for 2 minutes. Move the flow through to a new, clean eppendorf tube.
- Add 250 μl of clean 100% EtOH and mix by pipetting up and down. DO NOT VORTEX. A precipitate may form and that’s OK.
- Apply the solution (~600-700 μl) including any precipitate to an RNeasy column in 2 ml collection tube. The nucleic acids will bind the resin in these columns and the remaining cellular components will pass into the flow through.
- Spin the tubes for 30 seconds in a microfuge at 10,000 rpm. (THIS IS NOT FULL SPEED!!). Discard the flow through into the sink.
- Add 350 μl RW1 to the columns.
- Spin the tubes for 30 seconds at 10K (again NOT full speed!!). Discard the flow through into the sink AND the collection tube into the trash. Move the columns to clean 2 ml collection tubes.
- Add 80 μl of fresh DNaseI stock directly to the RNeasy membrane and incubate at room temperature at least 30 minutes.
- Add 350 μl RW1 to the columns and spin the tubes for 30 seconds at 10K. Discard the flow through into the sink and the collection tube into the trash. Move the columns to clean 2 ml collection tubes.
- Add 500 μl RPE to the columns.
- Spin the tubes for 2 minutes (!) at 10K (not full speed…last time). Discard the flow through AND the collection tubes. Move the columns to clean 2 ml collection tubes.
- Spin the columns to dry them for 1 minute, full speed in a microfuge.
- Trim the cap off two new 1.5 ml eppendorf tubes (save the caps!) and label the sides of the tubes with your team color, the date and a name for the sample. Transfer the columns into the trimmed eppendorf tubes and elute the RNA from the columns by adding 50 μl of RNase-free water to each.
- Spin the columns for 1 minute, 10K (oops, NOT full speed again!) then cap and store the samples on ice.
- Measure the concentration of your RNA sample by adding 5 μl to 495 μl sterile water. Use your P1000 to transfer the dilution to a quartz cuvette and measure the absorbance at 260 nm. Water in one of the optically paired cuvettes should be used to blank the spectrophotometer, but if another group has done this already, it does not have to be repeated.
- A few things to be aware of when using quartz cuvettes:
- They are very expensive.
- The lab has only one set.
- When you are done using the cuvette, you should carefully clean it by shaking out the contents into the sink and rinsing it two times with water. Quartz cuvettes get most of their chips and cracks when someone is shaking out the contents since it is so easy for the cuvette to slip from wet fingers or be hit against the sink. Don’t let this happen to you.
- A few things to be aware of when using quartz cuvettes:
- To determine the concentration of RNA in your sample, use the fact that 40 μg/ml of RNA will give a reading of 1 A260.
| RNA Sample | A260 | Conc of dilute RNA | Conc of undiluted RNA |
|---|---|---|---|
| Standard | |||
| Variable |
DONE!
For next time
- Calculate the number of cells in each microliter of your Lys-N-Go reactions. What assumptions are being used for converting the number of cells into the number of copies of LacZ DNA?
- Calculate the units of beta-galactosidase activity in each of the samples you measured.
- Calculate the concentration of RNA in each of your samples and determine the volume required for 1 ug of RNA.
- Continue to work on your writing assignment by finding a relevant historical example of technology with “dual-use” capabilities or societal pitfalls. For next time, please
- identify at least one reference that adequately describes the historical example you’ve chosen
- list the dual-use/societal issues related to the example you’ve chosen
- articulate however many similarities and differences you believe are relevant for this historical example and the emerging field of synthetic biology
- put on a black hat (figuratively speaking) and imagine then detail any dual-use capability for the system you are designing. Feel free to editorialize on these.
