20.109(S11):Prepare DNA for cloning (Day3)
Introduction
Last time you designed a modification to the Plux-λ promoter in order to weaken its basal expression in the absence of AHL — you hope. To test your idea, we will have to make a new DNA construct, pED-IPTG-YFD, where YFD is "your favourite design." Depending on how modest a change you made, there could be multiple ways to implement it; for example, a few DNA basepairs can readily be mutated by a PCR-like approach called site-directed mutagenesis. In our case, we will clone a modified Plux-λ insert into the pED-IPTG backbone. We are outsourcing insert construction to a company specializing in DNA synthesis (Integrated DNA Technologies, or IDT). Recall that you designed two strands, each with a restriction site overhang complementary to the cognate one on the backbone. Before the strands can be ligated into the backbone, they must be annealed together into double-stranded DNA. This can be done by heating the DNA above its melting temperature (94-95 °C) and then letting it slowly cool - very similar to how we denatured RNA and cooled it in Module 1, except this time we want two strands to anneal rather than to form independent secondary structures.
The backbone must also be prepared for cloning, first by restriction digest, and then by gel purification. (What could happen if we didn't purify the large backbone fragment of interest from the rest of the digestion mixture?) As you will see today, one of the enzymes that we are using for cloning is not very good at cutting plasmids. (Lesson: always read all the way through the product notes!) Thus, we have prepared pED-IPTG-INS for you that was digested overnight with XmaI and BamHI. Because this plasmid is large (a little over 10 Kbp), you will run it on a relatively low weight percent gel (0.7 %) and use a kit especially designed for long DNA to purify it. As with the kit we used in Module 1, silica particles are used in concert with a chaotropic salt and pH variation to bind, wash, and elute DNA in turn. This time the particles are loose rather than in a column format. While the gel runs, you will set up control digests to understand how they work and why we had to do this one for you in the interests of time.
Next time you will ligate the backbone and insert together, and then transform the new construct into bacteria. You will also continue gathering data about the 'broken' model system, namely the transfer function that maps IPTG concentration to Miller units. Combining this transfer function with the analogous one for the edge detector (that maps light intensity to Miller units) will allow us to relate the IPTG- and light-sensitive systems to each other. For the mapping to work well, you should treat all the different cell strains as consistently as possible. Thus, you will set up cells carrying the transfer function plasmid for overnight culture from an OD of 0.0025, just like you did for the complete IPTG-sensitive system.
Now is also a good time to add to our understanding of Plux-λ (specifically the lux part, as we covered λ last time) and PompC. In doing so, we will learn about two fascinating pieces of prokaryote biology: two-component signaling systems and quorum sensing.
Bacteria don’t exactly have a reputation as great communicators! However, it turns out that these single-celled organisms do ‘talk’ to teach other in primitive ways. One thing that is useful for a bacterium to ‘hear’ about is how large a community it is living in: this is called quorum sensing, and can help with resource management in a population of bacteria. In Vibrio fischeri the quorum sensing system is composed (in part) of luxR and luxI - as you may by now have guessed. The autoinducer is a homoserine lactone analagous to AHL (made by luxI), while luxR is the induced gene that in turn activates transcription of luminescence genes. The lux stands for light: V. fischeri glow (luminesce) when they reach a critical density, which happens only when they are growing in an associated host. This fascinating system is reviewed here while this paper focuses on illuminating the luxI promoter.
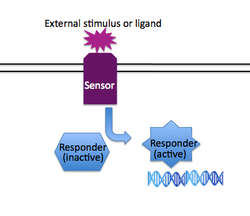
In addition to having group-based responses (such as quorum sensing), individual bacteria often must respond to changes in their immediate environment without regard for what other nearby bacteria are doing. Just as quorum sensing is a system that is implemented in different ways in many different species of bacteria, so is two-component signaling (2CS) a common solution to a type of problem faced by diverse bacteria . Generally speaking, two component systems consist of a "sensor" protein that spans the membrane, detecting an environmental cue on its exposed face, and transducing that information to the inside of the cell through a changed activity of its cytoplasmic face. The second component of traditional two component systems is a "responder" protein, usually a transcription factor that accepts the signal from the sensor protein and consequently regulates a set of genes to provide the cell with the proper response to the external stimuli. Examples of two component signaling systems include those that regulate chemotaxis, osmoregulation, and virulence. A review of 2CS from the perspective of appropriate signal transduction — e.g., limited cross-talk between different 2CS pathways — can be found here.
The relevant 2CS system in the edge detector is osmoregulation: the membrane-spanning protein EnvZ normally detects changes in salt concentration, phosphorylates itself and then transfers the phosphate to the responder protein OmpR. Phosphorylated OmpR can bind to both the OmpF and the OmpC promoters, and their differential affinities for OmpR-P help regulate appropriate porin expression on the cell membrane. In the edge detection (and bacterial photography) system, EnvZ is fused to the light-sensing protein Cph1, and native EnvZ is deleted from the bacterial strain used; thus, EnvZ autophosphorylation is regulated by light rather than salt concentration.
Protocols
A few notes before you begin
- The protocol parts are staggered today due to several incubation steps, and thus the day will not be as long (I hope!) as it may seem. Try to use your incubation times wisely. Parts 2-4 will require calculations.
- You will each have one large aliquot of nuclease-free water that you will need to use for several different protocols today. Be extra careful today about changing pipet tips, not dipping your pipet barrel into solutions, not pipetting so quickly that solutions shoot up into the barrel, etc. If at some point you think you need more clean water, don't hesitate to ask for it.
- Your DNA fragments for cloning will be stored at -20 °C until next time.
Part 1: Run digested backbone on gel
- Retrieve a 25 μL aliquot of digested backbone, and add 2.5 μL of loading dye to it.
- Load your entire sample onto the appropriate gel in the order listed below.
- We are leaving every other well blank both for ease of cutting out bands and so you are not all squeezing onto one gel.
- We will load the ladder.
- Once the gel starts running, continue with as many parts below as you can, and be sure to also pre-weigh an eppendorf tube for the purification (Part 5).
- When the gel stops running (45 min at 100 V), you will come and cut your bands out.
Gel 1:
| Lane | Sample | Lane | Sample |
|---|---|---|---|
| 1 | - | 6 | DNA ladder (load 10 μL) |
| 2 | Red group | 7 | - |
| 3 | - | 8 | Yellow group |
| 4 | Orange group | 9 | - |
| 5 | - | 10 | - |
Gel 2:
| Lane | Sample | Lane | Sample |
|---|---|---|---|
| 1 | - | 6 | DNA ladder (load 10 μL) |
| 2 | Green group | 7 | - |
| 3 | - | 8 | Pink group |
| 4 | Blue group | 9 | - |
| 5 | - | 10 | Purple group |
Part 2: Prepare short-term digests
Each lab team will prepare either a BamHI or an XmaI digest of the pED-IPTG-INS backbone. Although plasmid backbones must be cut with both enzymes before cloning (assuming two unique cut sites are used for that cloning), single-enzyme digests are important controls to run. With a short insert, long backbone, and low resolution gel, it is impossible to tell whether a plasmid has been cut once or twice. Either way the linear backbone will appear to be the same size.
The table below is for one reaction.
| Digest 1 (Red/Ylw/Blu/Prp) | Digest 2 (Org/Grn/Pnk) | |
|---|---|---|
| Plasmid DNA | 2.5 μL | 2.5 μL |
| 10X NEB buffer | 2.5 μL of buffer#4 | 2.5 μL of buffer#4 |
| BSA | 0.25 μL | 0.25 μL |
| Enzyme | 5 U of XmaI (50,000 U/mL stock) | 5 U of BamHI (20,000 U/mL stock) |
| H2O | For a total volume of 25 μL | For a total volume of 25 μL |
- Prepare a reaction cocktail for your assigned reaction above (digest 1 or digest 2) that includes water, buffer, BSA, and enzyme. Prepare enough cocktail so that you pipet no less than 1.5 μL of the limiting reagent, and then keep the cocktail on ice.
- Combine 2.5 μL of the backbone with 22.5 μL of the cocktail, incubate at 37°C for one hour, then move to ice.
- While your samples are digesting, you can continue with Parts 3-6 of the protocol.
- Before leaving lab today, please add 2.5 μL of loading dye to your digest. We will run them all on a 0.7% gel, along with ladder and undigested backbone for comparison, and post the image on the wiki.
Part 3: Prepare transfer function cultures
The cells you will use today are strain AB25, carrying plasmid pIPTG-lacZ. This plasmid omits the cell communication module from the pseudo-edge detection system.
- If you are waiting for the spectrophotometer, you can begin by picking up fourteen 14 mL round-bottom tubes and preparing sticky labels for your samples (described in step 6).
- Measure the OD600 value of a 1:10 dilution of your cells (use a total volume of 600 μL).
- In a 50 mL conical tube, prepare enough LB with ampicillin (stock is 1000X) for fourteen 2.5 mL cultures, plus 20% excess.
- Dilute the cell stock to an OD of 0.0025 in the LB-Amp solution.
- Distribute 2.5 mL of cells to each round-bottom tube with a 5 or 10 mL serological pipet.
- Ideally do this using the pipet-aid rather than a rubber bulb.
- Add IPTG to each tube in order to prepare duplicate samples with (0, 0.1, 0.25, 0.5, 1, 2.5, and 5 mM) IPTG.
- Recall that the IPTG stock is 1 M. The IPTG can be diluted in water.
- You may need to prepare intermediate dilutions of IPTG in order to avoid pipetting small volumes.
- Place your tubes on the roller wheel, remembering the following good practices:
- Completely snap shut the tube caps.
- Be careful not to tip the tube "backwards" as you insert it in the roller wheel.
- Balance your tubes as if on a centrifuge.
- Turn the roller wheel back on!
- Your cultures will be grown overnight, then moved to 4 °C until next time.
In order to most efficiently process a large sample number, you will all do a plate-based rather than cuvette-based Miller assay next time.
Part 4: Anneal oligos (if they have arrived)
- Resuspend each oligo at 100 μM in DNase-free water.
- In case any DNA got trapped in the cap during shipping, first do a short spin of the tubes.
- Prepare a mixture that has each oligo at 10 μM in ligation buffer in a total volume of 100 μL.
- When everyone is ready, the mixtures to be annealed will be placed on a heat block at 94-95 °C for 5 minutes, then slowly cooled by turning off the heat block but leaving the tubes inside.
- Be sure to write down the molecular weights of your oligos before leaving, as you will need these for your homework assignment.
Part 5: Purify digested backbone
- As you did in Module 1, cut the relevant band out of the gel with a clean spatula, while wearing both safety glasses and a UV-protective face shield.
- Keep your labmates aware of when the UV is on.
- Remember to avoid cutting out excess gel that does not contain DNA. Working with more than 250 mg of gel will be difficult.
- Get the weight of the gel slice by subtraction, then add the following: 3 volumes of buffer QX1, and 2 volumes of nuclease-free water.
- Vortex one of the QIAEX II bead aliquots for 30 seconds, then add 10 μL to your gel sample.
- Incubate for 10 minutes in the 50 °C water bath, flicking every 2-3 min to help dissolve the gel.
- If the color becomes orange or violet rather then yellow, see the teaching faculty. You will need to adjust the pH of your sample.
- Centrifuge for 1 min at maximum speed, and pull off the supernatant. Either balance your tube against another group's or use a water-filled tube.
- Throughout this protocol, you might initially use your P1000, then switch to the P200 to more easily pull off the last bit of liquid with a finer tip.
- Add 500 μL of buffer QX1, and flick the pellet to resuspend. Do not vortex, as this can shear long DNA fragments.
- Resuspension by flicking is a bit of an art, so feel free to ask the teaching faculty to demonstrate. With practice, you should be able to do it in about 30 seconds, not a few minutes.
- One method that works well is flicking while holding the tube upside down, then smacking it on the lab bench to return the partially resuspended pellet to the bottom.
- Centrifuge and remove the supernatant, then resuspend in 500 μL PE by flicking before centrifuging again.
- When you remove the PE after centrifugation, transfer it to a temporary waste container.
- Repeat the PE wash step with a fresh 500 μL of solution. This time, be careful to remove all of the supernatant or your pellet will not dry in time. Because the pellet is fairly stable, when it is almost dry you can turn the tube upside down and tap it to expel any fine droplets (smaller than you can see) of ethanol. Again, you can ask the teaching faculty for assistance if needed.
- The pellet should begin to turn white after 10 min, and be completely dry after 15-20 minutes.
- Note that the thin top smear will never turn white; if you wait for that, you will over-dry your pellet.
- Add 20 μL of EB buffer to the pellet and flick to resuspend. Due to the small volume, this resuspension step will take you longer to complete than the previous ones. Really tap the tube down hard on the bench to recover as much volume as possible (but do not centrifuge yet).
- To best dissolve this long fragment of DNA, put your sample in the 50 °C bath for 10 minutes, or at least for 5 min.
- Spin for 1 min, and then very carefully transfer the supernatant to a clean eppendorf tube. You should be able to recover 18-20 μL of fluid, but avoid the beads.
- To make sure that you have not taken up any beads, or to get them out of the way if you have, briefly (3-5 seconds) centrifuge your tube and look for formation of a pellet.
Part 6: Measure backbone recovery
- You will typically need 4-6 μL of backbone (total) for your control and experimental ligations. Being sure to reserve that much, measure the absorbance of your purified DNA.
- Use a minimum volume of 400 μL in the UV micro-cuvettes.
- To be above the detection limit of the spec., you will probably need to do an at most 1:50 dilution.
- Refer to the module 1 protocol if you are uncertain which absorbance readings you need to record.
- For DNA, an absorbance reading of 1 at the appropriate wavelength indicates a 50 μg/mL concentration (rather than 40 μg/mL as it was for RNA).
- A purity ratio near 2 is desired for both types of nucleic acids.
For next time
- Next time you will ligate together your backbone and unique insert. Calculate how much you need to use of each DNA based on the guidelines below.
- A good amount of backbone to use is 50-100 ng.
- It is typically helpful to have the insert slightly in excess, say a 2:1 molar ratio of insert to backbone.
- The size of the backbone is approximately 11,000 base-pairs. You can use an average per-base molecular weight to calculate the total molecular weight of this double-stranded DNA.
- The molecular weight of your oligo (before annealing) should already be written in your notebook based on the spec. sheets you received today.
- The total volume of DNA cannot be more than 9 μL for our reaction size.
- When the control digests are posted on the wiki, have a look at the gel image and answer the following questions.I forgot to lecture about this issue during pre-lab; please save this entire question until the D4 FNT.
- Does the uncut backbone run primarily in nicked or in supercoiled form?
- Which enzyme is a worse cutter, XmaI or BamHI?
- Look up the NEB notes page for the poor cutter. What protocol is recommended for plasmid DNA?
Reagent list
- Agarose gels (0.7%)
- Prepared in TAE buffer
- With 0.8 μg/mL ethidium bromide
- Loading dye
- 0.25% xylene cyanol
- 30% glycerol
- RNase
- Gels made and run in 1X TAE buffer
- 40 mM Tris
- 20 mM Acetic Acid
- 1 mM EDTA, pH 8.3
- 1 Kbp DNA ladder from New England BioLabs (NEB)
- Buffers and enzymes for restriction digest, as well as ligase buffer for oligo annealing from NEB
- LB and IPTG as on Day 1
- QIAEX II Gel Extraction Kit from Qiagen
