20.109(S09):Module 1
Module 1
Instructors: Alan Jasanoff and Agi Stachowiak
TA: Naiyan Chen
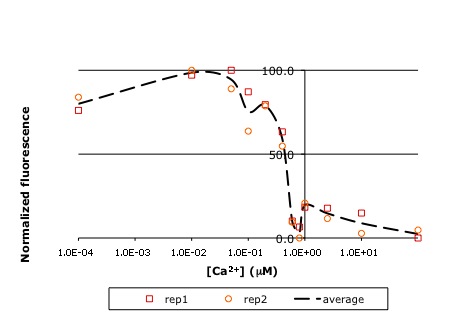
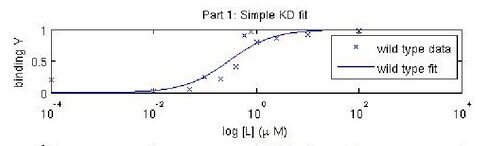
In this experiment, you will modify a protein called inverse pericam (developed by Nagai et al.) in order to affect its functions as a sensor. Inverse pericam (IPC) comprises a permuted fluorescent protein linked to a calcium sensor. The “inverse” in the name refers to the fact that this protein shines brightly in the absence of calcium, but dimly once calcium is added. The dissociation constant [math]\displaystyle{ K_D }[/math] of wild-type IPC with respect to calcium is reported to be 0.2 μM (see also figure below). Your goal will be to shift this titration curve or change its steepness by altering one of the calcium binding sites in IPC’s calcium sensor portion. You will modify inverse pericam at the gene level using a process called site-directed mutagenesis, express the resultant protein in a bacterial host, and finally purify your mutant protein and assay its calcium-binding activity via fluorescence. In the course of this module, we will consider the benefits and drawbacks of different approaches to protein design, and the types of scientific investigations and applications enabled by fluorescently tagged biological molecules.
We gratefully acknowledge 20.109 instructor Natalie Kuldell for helpful discussions during the development of this module, as well as for her prior work in developing a related module.
Module 1 Day 1: Start-up protein engineering
Module 1 Day 2: Site-directed mutagenesis
Module 1 Day 3: Bacterial amplification of DNA
Note: week off between day 3 and day 4 of lab.
Module 1 Day 4: Prepare expression system
Module 1 Day 5: Induce protein and evaluate DNA
Module 1 Day 6: Characterize protein expression
Module 1 Day 7: Assay protein behavior
Module 1 Day 8: Data analysis
