20.109(F08):Module 1
Module 1
Instructors: Bevin Engelward, Natalie Kuldell, Agi Stachowiak
In this experimental module you will modify the gene for EGFP (Enhanced Green Fluorescent Protein) to truncate the protein it encodes. Cells expressing the full-length protein glow green when exposed to light of the appropriate wavelength. You will be designing and then creating an expression vector to delete the first 32 amino acids of EGFP. Cells transfected with your expression vector should not glow green, a prediction you will test. You will also test whether this N-terminally truncated EGFP can recombine with a C-terminally truncated version to regenerate full length EGFP in vivo. Finally, you will have the opportunity to suggest changes to the experimental protocol that will increase the frequency of green cells in which there has been an inter-plasmid recombination event. We will then choose a few variables to test on the final day of the experiment.
Lablinks: day by day
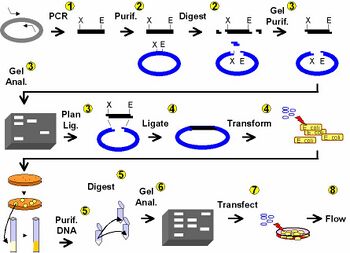
Day 1: DNA engineering using PCR
Day 2: Clean and cut DNA
Day 3: Agarose gel electrophoresis
Day 4: DNA ligation and bacterial transformation
Day 5: Examine candidate clones
Day 6: Restriction map and tissue culture
Day 7: Lipofection
Day 8: FACS analysis
DNA engineering lab report guidelines
DNA engineering P3 guidelines
References
- DNA double-strand break repair: From mechanistic understanding to cancer treatment
DNA Repair 2007
Thomas Helleday, Justin Lo, Dik C. van Gent, Bevin P. Engelward
URL
Sample Animation Animations were made by Justin Lo (BE class of '08), a former UROP student in Professor Engelward's laboratory! - Homologous recombination as a mechanism of carcinogenesis
Biochim Biophys Acta 21 March 2001
Bishop AJ and Schiestl RH
URL - Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death
EMBO J 15 January 1998
E Sonoda, M S Sasaki, J M Buerstedde, O Bezzubova, A Shinohara, H Ogawa, M Takata, Y Yamaguchi-Iwai, and S Takeda M
URL
