IGEM:University of Chicago/2009/Notebook
Projects
- Biosensor
- PNP biosensor
- Organophosphate Hydrolase expression
- PNP degradation
- Human Practices
- Standards
Project Summary
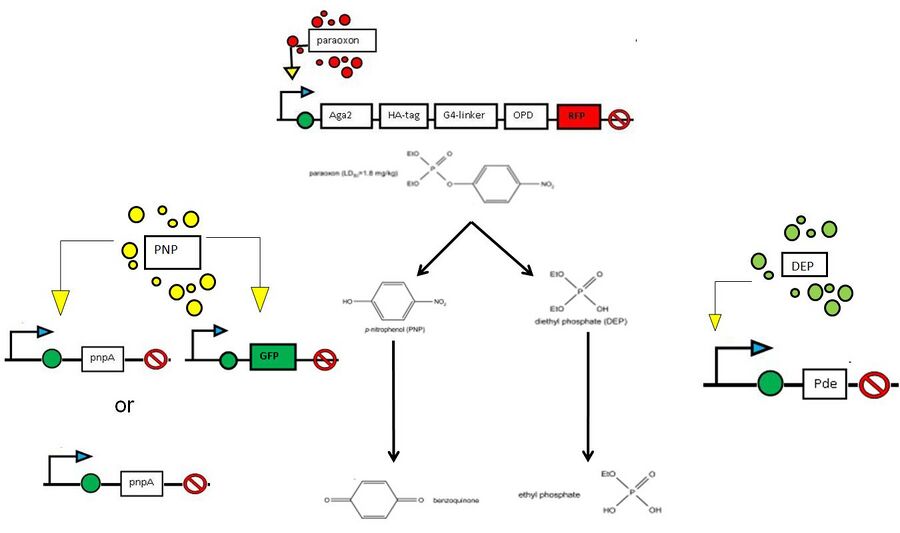
Note: This project summary reflects our initial plan, which involved paraoxon,PNP,or DEP inducible expression of degradation genes. Because our genes arrived late, and given the relative lack of sensitivity of our promoters to the above compounds, we decided to constitutively express degradation genes under ADH1 constitutive promoter.
Who are we? In short, we are the University of Chicago iGEM team (and Synthetic Biology Registered Student Organization). Our project this year is (officially) "A yest-based biodegradation and biosensor system for the organophosphate neurotoxin paraoxon." While labs in the past have worked with similar systems and produced excellent results (Keasling Lab at Berkeley and Mulchandani Lab at UC Riverside for example) we've decided to do things a little differently. Using yeast as our model organism, we hope to create an efficient degradation and biosensor package--that is, yeast which will both break down paraoxon and mark the progress of degradation. We invite you to keep up with our projects, and we'll do our best to update these notebooks as often as possible. Enjoy!
Why paraoxon?
Paraoxon is the active metabolite of the commercial pesticide parathion. It is part of a family of organophosphate neurotoxins that include chemical warefare agents such as VX, sarin, and soman. All these compounds are acetylcholinesterase inhibitors--they work by inhibiting the breakdown of acetylcholine by the cholinesterase enzyme, causing prolonged muscle contraction, respiratory depression, and in fatal cases, asphyxiation.
Why yeast?
- Saccharomyces cerevisiae, or common bakers yeast, has number of benefits over bacteria (both E. coli and Pseudomonas Putida have been used for paraoxon degradation in the past). First off, most genetic variants have been characterized and are easily accessed. Yeast is also more easily lyophilized and stored, and yeast genotoxic-inducible genes respond to a much broader spectrum of damaging agents than bacteria.
Paraoxon biosensor
- For our first project, we hope to create a straightforward paraoxon biosensor through chromosomal integration of GFP or RFP downstream of a series of paraoxon-sensitive promoters identified by Schofield et al (Appl Microbiol Biotechnol. 2007 Oct;76(6):1383-94). The genes for these promoters were up-regulated up to 1,700 fold in the presence of paraoxon. In addition to being permanently incorporated, unlike a plasmid which needs constant selection in order to be retained within the cell, these promoters can be used for endogenous expression of Organophosphate hydrolase and/or subsequent degradation genes.
PNP biosensor
- Our second biosensor utilizes another set of genes (also identified by Schofield et al) that are sensitive to paraoxon hydrolysis. That is, why yeast cells were grown which expressed organophosphate hydrolase, these genes were up-regulated. between 2 and 6-fold. None of these promoters have been identified as PNP-sensitive per-se (P-nitrophenol, a byproduct of paraoxon degradation), so part of our task will be identifying which promoters are indeed PNP sensitive. As with the paraoxon-sensitive promoters may be used for expression of degradation genes.
Organophosphate hydrolase surface expression
- For this project, we hope to efficiently express organophosphate hydrolase, a paraoxon degrating enzyme endogenous in a number of soil bacteria such as Flavobacterium, in yeast (Saccaromyces Cerevisiae).
PNP degradation
- Our final (and most difficult task) will be to create a workable PNP, and thus paraoxon, degradation system in yeast. We hope to integrate degradation genes taken from Pseudomonas ENV2030 to break down P-nitrophenol into a usuable carbon source. We also hope to use the second by-product, diethyl phosphate, as a phosphate source.
Human Practices
- In addition to our competition project, our human practices division (in conjunction with University of Illinois at Urbana-Champaign) is planning a series of genetic engineering presentations at local schools. These presentations will be construction from a "kit" of interchangeable slides and topics we will design, and can be used by later teams for their own similar presentations.