Harmer Lab:Research
|
Room 2123 |
BackgroundAs one adage has it, the only constant is change. A striking example of such constant change is the regular alterations in the environment caused by the daily rotation of the earth on its axis. Along with the obvious diurnal changes in light and temperature, other important environmental variables such as humidity also change on a daily basis. This periodicity in the geophysical world is mirrored by daily periodicity in the behavior and physiology of most organisms. Examples include sleep/wake cycles in animals, developmental transitions in filamentous fungi, the incidence of heart attacks in humans, and changes in organ position in plants. Many of these daily biological rhythms are controlled by the circadian clock, an internal timer or oscillator that keeps approximately 24-hour time. Less obviously, the circadian clock is also important for processes that occur seasonally, including flowering in plants, hibernation in mammals, and long-distance migration in butterflies. In fact, circadian clocks have been found in most organisms that have been appropriately investigated, ranging from photosynthetic bacteria to trees [1]. 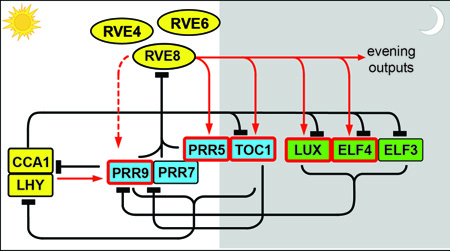 Circadian rhythms have been studied for hundreds of years, with plants used as the first model system (see [3] for an excellent summary of the history of clock research in plants). As rooted organisms living in a continually changing world, plants are masters at withstanding environmental variation. The circadian clock is key: it both ensures the optimal timing of daily and seasonal events to cope with predictable stresses and regulates myriad signaling pathways to optimize responses to environmental cues. Perhaps for these reasons, the transcriptional network underlying the plant circadian oscillator is uniquely complex and a higher percentage of the transcriptome is under clock control than in other eukaryotes. In addition to regulated transcription, post-transcriptional regulation is also clearly essential for proper clock function. Our lab is interested in understanding the molecular nature of the plant circadian clock and how the clock influences plant physiology.
Understanding the plant circadian oscillator
 Like their well-known homologs CCA1 and LHY (reviewed in [6]), the RVEs bind to evening element (EE; AAAATATCT) sequences in the regulatory regions of both central clock genes and output genes. However, while the RVEs activate expression of evening-phased target genes, CCA1 and LHY repress expression of these same genes. Thus these related proteins carry out antagonistic biochemical functions within the circadian clock. We are currently investigating the mechanism by which these Myb-like transcription factors control rhythmic expression of clock and output genes. In related work, we are investigating the role of a novel gene in the plant circadian clock. We identified XCT in a forward genetic screen for period mutants using a luciferase reporter gene (Figure 2) [7]. Luciferase imaging is a relatively high throughput, non-destructive, and automated way to monitor circadian rhythms in plants. Later, we discovered that XCT is also involved in ethylene signaling [8]. XCT is highly conserved across eukaryotes; not only can the Arabidospsis gene complement fission yeast mutant for the XCT ortholog Xap5 [9], but Xap5 can rescue xct mutant plants (Figure 3).  Genetic, transcriptomic, and chromatin immunoprecipitation data suggest that yeast Xap5 acts in a similar manner as the variant histone H2A.Z to regulate gene expression and suppress expression of aberrant transcripts[9]. We are now investigating whether XCT plays a similar role in Arabidopsis and working to identify a biochemical function for the XCT protein. Clock regulation of physiologyPlants with a circadian period matching that of environmental light/dark cycles have a growth advantage [10]. It is therefore perhaps not surprising that most species exhibit daily rhythms in leaf, stem, and root growth [11]. Growth rhythms may be driven directly by environmental factors such as light, temperature, and water availability, but many are the product of complex interactions between external cues and the circadian system[12, 13, 14, 15]. Although great advances have been made in understanding how the clock and environment cooperate to control growth, most such studies have been carried out using the embryonic stem (or hypocotyl) of Arabidopsis as a model. Investigations using different systems are likely to provide novel insights into how plant growth is optimized with the environment.  To better understand links between the clock, environmental sensing pathways, and growth regulation, we are now collaborating with the Blackman Lab to study growth regulation in sunflower. Sunflower is well-known for diaheliotropism or solar tracking, with plants changing the angle of their leaves and stems over the day so that they remain perpendicular to the sun’s rays. This special type of rhythmic growth is found in many plants (including various crop and pasture species) and is associated with increased yield, likely due to increases in photosynthetic and water use efficiency [16, 17, 18]. The conspicuous movement of aerial organs from east to west during the day is likely mediated by activity of the phototropin blue light photoreceptors [19]. Much more mysterious is the reorientation that occurs during the night so that sunflower leaves and apices face east well before the sun rises (Figure 4) (see also Plants-In-Motion for a wonderful movie of a solar-tracking sunflower).
Thus solar tracking is in part a phototropic response, but it also entails directional growth at night in anticipation of the timing and direction of dawn. This anticipatory behavior suggests involvement of the circadian clock. We have confirmed this in growth experiments carried out in environmental control chambers, and are now taking genomic and metabolomics approaches to identify genes and pathways involved in solar tracking. In the future, we plan to carry out functional analysis of selected candidate genes and pathways, taking advantage of the genomic tools now becoming available in sunflower. Exploration of the links between the clock and diverse signaling pathways will lead to a better understanding of how the circadian clock affects plant growth and leads to improved fitness.
References
|
