Biomod/2013/Harvard/design
<html>
<head>
<link href='http://fonts.googleapis.com/css?family=Open+Sans' rel='stylesheet' type='text/css'>
</head>
<style>
body {
font-family: 'Open Sans', sans-serif; overflow-y: scroll;
}
.container {
background-color: #ffffff; margin-top:0px
} .OWWNBcpCurrentDateFilled { display: none; }
h1 {
font-size: 36px; line-height: 36px; padding-top: 5px; border-bottom-width: 0;
}
h3 {
font-size: 18px;
}
h5
{
font-family: 'Open Sans', sans-serif; font-size: 11px; font-style: normal; text-align: center; margin:0px; padding:0px;
}
- column-content
{
/* Uncomment to Dewikify width: 0px; float: left; */ margin: 0 0 0 0; padding: 0;
} .firstHeading {
display:none; width:0px;
}
- column-one
{
display:none; width:0px; padding-top: 35px; background-color: #ffffff;
}
- globalWrapper
{
width: 900px; background-color: #ffffff; margin-left: auto; margin-right: auto
}
- content
{
margin-left: 0px; margin-top: 0px; padding-top: 0px; align: center; /*padding: 12px 12px 12px 12px; width: 30%; background-color: #ffffff; border: 0; */
}
- bodyContent
{
width: 850px; align: center; background-color: #fffffff;
}
- column-content
{
width: 900px; background-color: #ffffff;
}
- footer
{
position: center; width: 900px;
} @media screen {
body { background: #000000 0 0 no-repeat; /* changed default background */ }
}
- menu
{
align: left; width: 10em; padding: 0px 10px 10px 10px; background-color: #FFFFFF; float: left;
}
- pagecontent
{
width: 620px; min-height: 400px; float: left; margin-left: 0px;
}
.group:after {
content: ""; display: table; clear: both;
}
.editsection {
/*display: none*/
}
a:link {color:#FF6060;}
a:active {color:#B24343; }
a:hover {color:#B24343; text-decoration: none}
a:visited {color:#FF6060;} /* visited link */
/*Expanding list*/
- exp { list-style: none; }
- exp li {
height: 1.8em; border-top: 1px solid #dedede; margin: 0 0 0 0; padding-top: .2em
}
- exp li:hover { background-color: #F8F8F8}
- exp li a:hover { display: block }
</style> </html>
Design
The modular approach naturally divides the project into two parts: the input domain and output domain.
Input
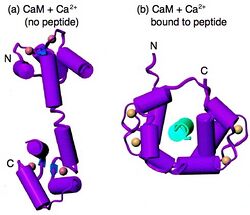
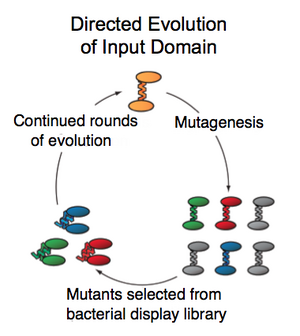
Directed evolution was performed by screening individual purified members of the library for activity toward a previously inactive peptide, Staphylococcus aureusδ-toxin. The Calmodulin portion of the BlaCaM fusion gene was mutagenized randomly using error-prone PCR and cells expressing those mutants were grown in 96-well plates. The expressed BlaCaM library members were purified via a 6xHis tag and screened for δ-toxin-activated β-lactamase activity in vitro. Members showing increased δ-toxin activity relative to a negative and positive control were re-screened to verify, and then combined and mutagenized again to continue a second generation of the evolution. After as many rounds of diversification and screening as possible, the selected library members will exhibit binding to a previously unrecognized peptide.
Output
Overview
The output team's goal is to alter the BlaCaM switch to produce a different output signal by replacing the transducer region of BlaCaM (split beta-lactamase) with another protein that will produce a different output signal in response to the conformation change of CaM. Among various forms of output signals--fluorescence, luminescence, electrical signal, enzymatic activity, etc.-- we chose luminescence for the relative ease in detection/visualization of luminescence, and furthermore, we chose to engineer Guassia princeps-derived luciferase (GLuc) as the output domain protein to replace the beta-lactamase region of the existent BlaCaM.
GLuc and GLucCaM
GLuc catalyzes the oxidation of the substrate coelenterazine to produce light of peak wavelength length around 480nm, as shown in the figure below:
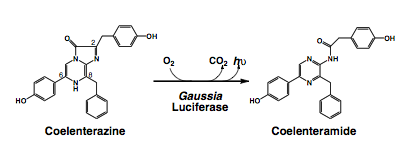
Many features of GLuc distinguished GLuc as a highly attractive output domain for the project. GLuc has a strong activity with its luminescence intensity upto 700-fold higher than that of Renilla or firefly-derived luciferases; it is highly stable, as it remains fully folded up to 40˚C and even retains 65% of its activity after 30 minutes of incubation at 95˚C; lastly, it is one of the smallest luciferase (20kD).
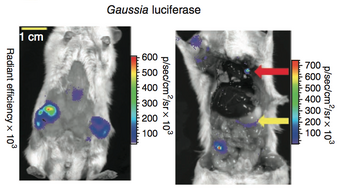
Moreover, GLuc is already gaining popularity as a reporter gene. For example, the following picture shows a in vivo imaging using GLuc as the reporter.

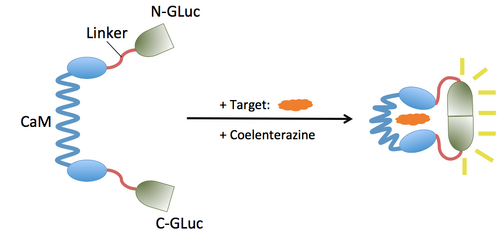
Our goal was to replace the beta-lactamase output of BlaCaM with the GLuc output while keeping the CaM functionality intact. To do so we replaced the beta-lactamase regions of BlaCaM with split GLuc to produce GLucCaM, a protein switch that activates GLuc activity in response to conformation change of CaM upon target binding. Specifically, we sought to:
- Create a GlucCaM that retains the chemiluminescence function of GLuc.
- Explore different structure-activity relationships in GlucCaM
- Engineer GLucCaM to output chemiluminescence specifically in response to the target binding conformation changes of CaM.
Design of GLucCaM
GLuc will be split into two, yielding us the N-terminal and C-terminal halves of Gluc (N-GLuc and C-Gluc). These two will replace the existing N-terminal and C-terminal halves of beta-lactamase in BLaCaM. We desire the GLucCaM switch to deactivate when CaM binds to Ca(2+) and assumes a rigid shape that separates the two halves of GLuc by distance, and then upon target binding, it should activate GLuc as CaM's conformation change brings the two halves into proximity.
The first task is retaining the functionality of GLuc through the splitting. The next task is to vary many parameters to test the function-structure relationship of the switch. This step includes varying linker length, linker rigidity, mutations on Cysteine residues, expression conditions, etc. Some possible concrete challenges of this step are: 1) linkers may be too short or long, in which case the GLuc may never be active or constitutively active, 2) 10 disulfide bridges present in GLuc may prevent the two halves of GLuc from separating once they are brought to proximity, and 3) it has been noted that E.coli may not be the optimal expression medium for GLuc.
The schematics of what we aim our final product to be be is as follows: