BISC219/F13: Lab 5
Lab 5: Continue Mapping the Mutation
Because Fall Break is coming and we do not want you to have to come in to do work during break, please place 2 young adult double mutant hermaphrodites from a true breeding plate onto new each of 2 new plates. Make sure there are no males on the plate from which you select your worms! Label these plates with your PURPLE Sharpie.
We will incubate your worms at lower temperature to slow them down until Lab 6.
Complete Complementation Testing
Examine your complementation crosses. The main consideration is whether or not there are any MALE progeny of Dpy phenotype present in the progeny of cross #2. WHY male? Think about the hallmark of a successful cross! What else can hermaphrodites do other than mate with males - might be why you have Dpys on more than one plate.
If you find a cross with Dpy male progeny pick one or two to a fresh plate and take their picture being sure to get their tails in the picture. This is your "proof" of failure to complement.
For each plate consider that:
A) If your unknown mutation and your known mutation under study are in different genes (not allelic): the heterozygote male that you incorporated into Cross #2 will produce two types of gametes: type 1) with a dpy-u mutation and a wild type copy of the dpy-k gene and type 2) with wild type dpy-u gene and wild type dpy-k gene. One the other hand, the hermaphrodite you added will produce only one type of gamete-- we'll call it gamete type 3) with wild type dpy-u gene and a mutation in dpy-k.
These gametes can be combined so that you will have two possible genotypes. Combining gametes type 1 and 3 you get dpy-u from the male and wild type for the hermaphrodite so the progeny are heterozygous at the dpy-u locus (dpy-u/+) and for the dpy-k locus you get wild type from the male and dpy-k from the hermaphrodite (+/dpy-k). These individuals are phenotypically wild type since they have one wild type allele for each of the genes. Combining gametes 2 and 3 for the dpy-u locus you get wild type alleles from the male and the hermaphrodite (+/+) and for dpy-k you get wild type from the male and dpy-k from the hermaphrodite (+/dpy-k). These individuals are phenotypically wild type since they have at least one wild type allele for each of the genes. In conclusion if your unknown mutation (dpy-u) and your known mutation are in different genes (not allelic) will you observe any mutants in the progeny of cross #2?

B) If your unknown mutation (dpy-u) and one of the previously identified mutations under study (dpy-k) are in the same gene (allelic): the heterozygote male that you incorporated into Cross #2 will produce two types of gametes: type 1) with a dpy-u mutation and type 2) with wild type gene. One the other hand, the hermaphrodite you added will produce only one type of gamete we'll call it gamete type 3) with the dpy-k mutation.
These gametes can be combined so that you will have two possible genotypes. Combining gametes type 1 and 3 you get dpy-u from the male and dpy-k from the hermaphrodite so the genotype is (dpy-u/dpy-k). These individuals are phenotypically mutant. Combining gametes 2 and 3 you get wild type from the male and dpy-k from the hermaphrodite so the genotype is (+/dpy-k). These individuals are phenotypically wild type since they have one wild type allele. In conclusion if your unknown mutation (dpy-u) and your known mutation are in the same gene (allelic) will you observe mutants in the progeny of cross #2
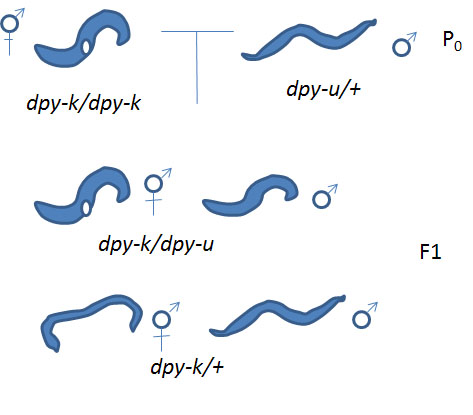
Can you now identify the dpy gene responsible for your Dpy phenotype?
If you discovered that one of the reference 'dpy strains was allelic to your dpy mutation, congratulations! You now know the name of your mutated gene and are ready to start the next phase of our Forward Genetics study: characterization of the nature of the mutation and, perhaps, discovery of a functional area of the protein product. DNA sequence analysis and bioinformatics are next.
If none of your reference strains failed to complement (meaning that everything tested was non-allelic), you may not have learned much about the identity of the gene associated with your mutation but that is not, necessarily, a bad thing. Does that make it more likely that the gene you are characterizing has not been previously identified as associated with a Dpy phenotype and may be a newly characterized mutation? It will be a lot easier to publish this work if so:)
Complementation Testing Analysis
From your complementation analysis, you may have discovered that the mutation you are studying is likely to have been previously characterized. If one of the known dpy mutants was shown to be allelic to your unknown dpy mutant, you may be able to find out from this complementation analysis the name of your dpy gene of interest. To learn more, you can enter the dpy gene name of the allelic reference strain into Wormbase.
Click on the link to Wormbase above and enter the gene name into the box at the top of the page and click Search. It will either bring you directly to that page or it will bring you to a page with multiple hits - click on the link that provides a definition for what the gene does.
On this new page you should find all the known information about this particular gene: its name, who named it, what the gene encodes - if that is known, and much more. At the bottom will be a list of references - or a link to a list of references. Does it appear that you are working with a well characterized gene? Do not be too disappointed if this is so. There are likely to be several different functionally significant areas on the gene that is associated with your mutation. Our mapping and DNA sequencing work will tell you more about the mutation and may help you pinpoint a functionally significant part of the gene.
Spend some time with Wormbase and marvel at all the hard work and years of research that went into discovering all this information about this tiny little nematode that causes us no harm (non-parasitic). Why do you think so many smart people have devoted so much of their time and energy to working out the genetics of "appearance or movement challenged" little worms? We will talk more about model organisms and the power of functional and comparative genomics in our next series.
Assignment
There is no graded assignment due in the next lab:)
