Wintrode:Research
Serpins: Folding, Misfolding and Function.
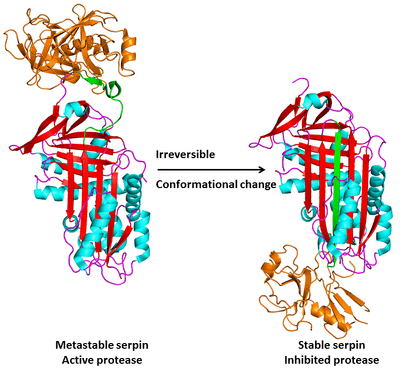
The serpins are an unusual class of serine and cysteine protease inhibitors that inhibit their target proteases through a unique mechanism. Serpins initially fold to a metastable structure and subsequently undergo a massive conformational change to a stable structure when they inhibit their target proteases. When the protease (orange) cleaves this reactive center loop (RCL, shown in green), the reaction proceeds through the formation of an intermediate in which the protease and the serpin are covalently linked. Before the breakdown of the intermediate brings the reaction to completion the RCL inserts into the central β-sheet (β-sheet A) and becomes a sixth strand, in the process translocating the target protease to the opposite end of the serpin molecule. The protease active site is distorted in this process, leaving it irreversibly trapped in a protease-serpin complex.
This "molecular mousetrap" inhibitory mechanism raises a number of questions. How do serpins get trapped in a metastable conformation during folding? How is the energy required for translocation and inhibition of the target protease stored in the metastable structure and how is the energy utilized during inhibition? The strained, metastable nature of serpin's native structure leaves them highly susceptible to disruption by mutations. A class of genetic disorders known as the serpinopathies have as their basis the propensity of mutant serpins to misfold and polymerize, resulting in decreased levels of secreted serpins and, ultimately, cell death when they accumulate in the ER. We are actively studying two specific serpinopathies: 1) alpha-1antitrypsin deficiency, which results from the misfolding of the serpin alpha-1 antitrypsin and leads to both liver disease and emphysema, and 2) Familial encephalopathy with neuroserpin inclusion bodies (FENIB), which results from the misfolding of neuroserpin and leads to early onset dementia.
We are using chemical labeling with mass spectrometry, fluorescence spectroscopy, and electron paramagnetic resonance spectroscopy to probe the folding and misfolding pathways of serpins as well as the structure of pathogenic misfolded serpin polymers. More information on the serpins can be found here.
Statistical Mechanics of Protein Folding and Conformational Change
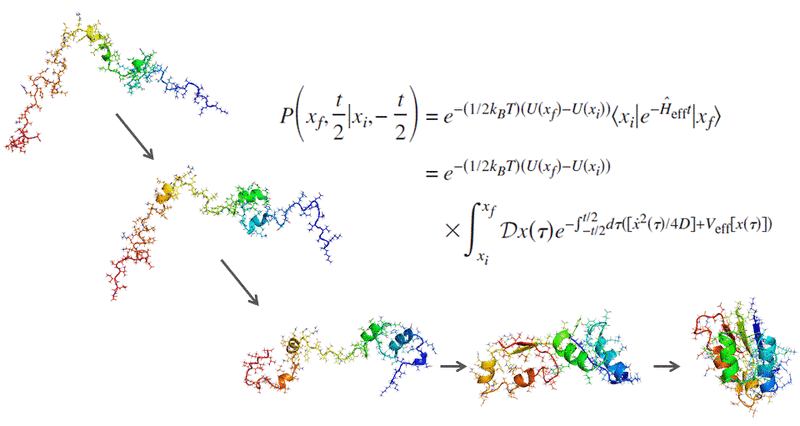
In collaboration with Professor Pietro Faccioli's group at the University of Milan, we are applying the recently developed Bias Functional (BF) method to simulate protein folding and conformational change. The extreme computational efficiency of the BF method allows us to avoid coarse graining and simulate systems in all atom detail using realistic physics based force fields. As a result, we are currently performing all atom simulations of conformational changes, folding and misfolding in systems of unprecedented size and complexity.
Modeling Protein Ensembles with Hydrogen/Deuterium Exchange and Multiscale Simulations
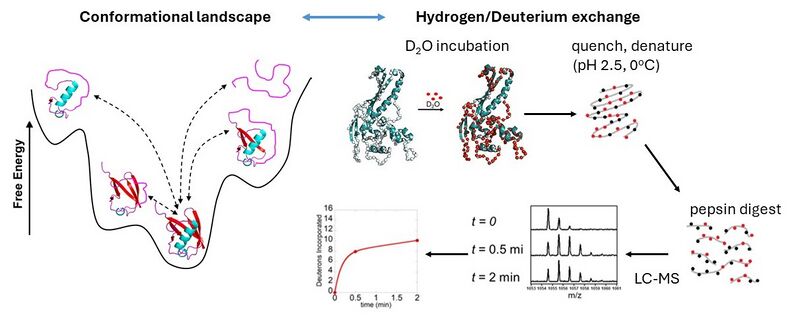
Proteins exists in solution as dynamic ensembles of rapidly interconverting structures. A premier experimental technique for characterizing protein dynamics is hydrogen/deuterium exchange (HDX). When a protein is immersed in D2O, hydrogen atoms along the backbone will exchange with deuterium, and the rate of this exchange is determined by local structure and structural fluctuations. HDX thus monitors structure and dynamics across the entire protein in a single experiment. Making full use of the information contained in HDX data requires an atomistically detailed molecular picture, which HDX experiments do not provide. Molecular dynamics simulations provide an atomistically detailed picture of protein dynamics, but their ability to explore conformational space is restricted by the limited timescales (microseconds) that are accessible for most proteins. One approach to overcoming this limitation is multiscale simulations, in which a coarse grained representation of the protein is used to rapidly explore conformational space, followed by reconstruction of an atomistically detailed ensemble. We are currently exploring the potential of machine learning methods to integrate multiscale simulations with experimental HDX data in order to rapidly generate realistic representations of protein structural ensembles.