Wet lab work-Glutamate
QBP PCR (Successful) - March 1, 2012 (1-3)
Goal:
Run a successful PCR with QBP.
Phusion PCR Reagents (50 μL total):
- ddWater (35μL)
- 5x Phusion Buffer (10μL)
- 10 mM dNTPs (1.5μL)
- Forward Primer BI141 row 37
(1μL)
- Reverse Primer BI142 (1μL) row 38
- diluted template DNA (E.coli) (1μL)
- Phusion Polymerase (.5μL)
Phusion PCR Protocol:
We mixed all the above reagents except for the E.coli template and the polymerase. We prepared the template by boiling the E.coli. We extracted some of the water and used that as the DNA template. Once we had the template we added it and the polymerase to the test tube. Then we placed it in the PCR machine and ran the PCR program.
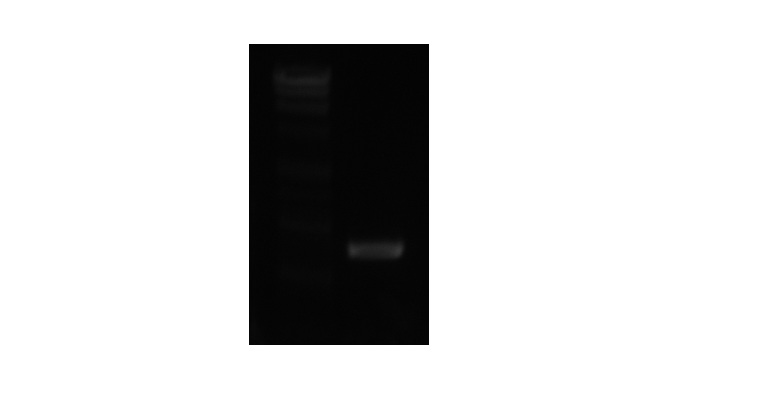
Results:
We did the above protocol and ran out gel and had a band at the appropriate place.
Purifying Our PCR Product (QBP forward & reverse)- March 2, 2012 (2-1)
Goal:
Successfully purify our PCR product.
Protocol:
1. Add 5 volumes of Buffer PB to 1 volume of the PCR reaction and mix.
2. Place a QIAquick column (referred to as "column" for now on) in a provided 2 mL collection tube.
3. To bind DNA, apply the sample to the column and centrifuge for 30-60 seconds. Discard flow-through and place the column back in the same tube.
4. To wash, add .75mL (750 μL) Buffer PE to the column centrifuge for 30-60 seconds. Discard flow-through.
5. Centrifuge the column once more in the provided 2 mL collection tube for 1 min to remove residual wash buffer.
6. Place each column in a clean 1.5 mL micro-centrifuge tube.
7. To elute DNA, add 50 μL Buffer EB (10mM Tris Cl, pH 8.5) or water (pH 7.0-8.5) to the center of the column's membrane and centrifuge the column for 1 min. For increased DNA concentration, add 30μL elution buffer to the center of the column's membrane, let the column stand for 1 min, and then centrifuge.
8. If the purified DNA is to be analyzed on a gel, add 1 volume of Loading Dye to 5 volumes of purified DNA. Mix the solution by pipetting up and down before loading the gel.
-QIAGEN PCR purification Kit (50)
Results:
After completing the above protocol we ran the product out on agarose gel and had a correct band.
Restriction Digest of PCR product (QBP) - March 6, 2012 (2-2)
Goal:
To use restriction digest on our PCR product.
Reagents and Protocol:
- PCR product (41.5μL)
- 10x NEB Buffer #3 (5μL) BR
- 100x BSA (0.5μL)
- PSTI (1.5μL)
- XhoI (1.5μL)
(Total 50μL)
1. Mix all together and put in water bath for 2 hours at 37°C.
Restriction Digest of Plasmid(pIG12) - March 8, 2012 (3-1)
Goal:
Digest the plasmid.
Reagents and Protocol:
- Plasmid (pIG12) (20μL)
- ddWater (21.5μL)
- 100x BSA (0.5μL)
- 10x NEB buffer (5μL)
- PSTI (1.5μL)
- xhoI (1.5μL)
(Total 50μL)
1. Mix well.
2. Let sit for at least 1.5 hours.
Low-Melt Gel - March 9, 2012 (3-1 and 3-2)
Goal:
To run out our digested PCR Product (QBP) and our digested plasmid (pIG12) on a low-melt gel, and then to cut those DNA bands out and store them in separate tubes.
Results:
Successfully ran the gel and got bands of the expected length (600-700 bp). We cut out the bands and added them to individual tubes .
-QBP Purified Restriction (3-2)
Ligation of digested Plasmid and digested PCR product - March 13, 2012 (3-3)
Goal:
Ligate the digested plasmid with the PCR product.
Reagents and Protocol:
1. Melt Gel at 65°C (QBP 3-2)
- Water (6.5μL)
- 10x Ligase Buffer with ATP (1.5μL)
- T4 DNA Ligase (1.0μL)
- pIG12 (3μL)
- insert (digested QBP) (3.0μL)
(Control does not include the above insert)
2. Incubate at room temperature for at least 30 minutes.
Transformation of Ligated Plasmid into DH5α E. coli cells - March 15, 2012 (4-1)
Goal:
Get the E. coli to uptake our plasmid.
Reagents and Protocol:
1. When the DH5α is thawed, quickly add 2.0μL of the ligation mix (melt ligation at 65°C) to 25μL of competent cells.
2. Vortex the tubes briefly and put them back in the ice for 5-10 mins. (keep DH5α as cold as possible during this process)
3. Heat shock @ 42°C for 60 seconds and immediately place the tubes back on ice for 2-5 mins.
4. Add 0.5mL of plain LB to the reactions and incubate at 37°C for 30 mins (because it uses ampicillin selection)
5. Plate 100μL of cells and incubate at 37°C over night.
PCR of Colonies - March 16, 2012 (4-2)
Goal:
PCR the DNA of the successful colonies.
Reagents and Protocol:
- Add one colony to 50 μL of water in a PCR tube. (We had 8 colonies [a-h])
- Streak diluted colonies onto a new plate (one plate with 8 sections)
- Boil PCR tubes (4-2 A) for 5 min in the PCR machine to release DNA.
- Set up Taq PCR rxns: (4-2B)
1. Water (19μL.)
2. Standard 10x rxn buffer (2.5μL)
3. 10mM dNTPs (0.5μL)
4. Each primer (IG11, IG12) (0.5μL)
5. Taq DNA polymerase (0.5μL)
6. Boiled colony sample (2μL)
- Place all reagents in PCR machine for at least 25 cycles.
Ran gel of Colonies PCR - March 20, 2012 (5-1)
Goal:
To compare bands and find which colonies have the approriate DNA band.
Results:
No bands appear out side of the wells. Only the loading dye

Plasmid Digest - March 22, 2012 (5-2)
We followed the protocol found here. We are going to run PCR on QBP again to try and get better results.
Restricted Plasmid tube: 5-2A
PCR QBP (5-3A) - March 23, 2012
We decided that it would be better to remake all of our products. We followed the same protocol as before.
We will most likely run the gel on monday.
Tubes:
Template DNA from PIG 125 (5-3A)
PCR QBP product (5-3B)
Gel of PCR Product (5-3B) - March 27, 2012
We ran the gel with our PCR product (5-3B) and got a band between 500- 800 base pairs, which is exactly where we wanted it to be.
Purification of PCR Products (5-4) - April 24-27, 2012
April 24, 2012
We followed the protocol found here. We also prepared a gel to run out our purified QBP PCR product (5-4A). JM
April 25, 2012
We ran our PCR purification (5-4A) out on gel to check that we had the correct number of base pairs (around 700 bps) for QBP. We only had a smudge, not a full band, so we need to run our purified PCR product (5-4A) again on the gel. Dr. Grose said the smudge could have been caused by a defect in lane 2. BR
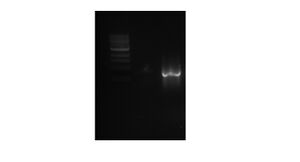
JA
April 27, 2012
We set up another gel to run our purified PCR product (5-4A) again to see if it was a defective lane, or if our product is not correct. A band appeared in the correct place, so we'll move on to the restriction digest of our purified PCR product. BR

JA
Restriction Digest of PCR product (6-1) - April 30, 2012
We followed the same protocol found here. BR
Low-Melt Gel - May 1, 2012 (6-2)
Goal:
To run out our digested PCR Product (6-1) and our digested plasmid (5-2A) on a low-melt gel, and then to cut those DNA bands out and store them in separate tubes.
JA
Results:
Successfully ran the gel and got bands of the expected length for QBP (600-700 bps) and pIG12. We cut out the bands and added them to individual tubes (QBP in 6-1 and pIG12 in 5-2A). BR
Ligation of digested plasmid and digested PCR product - May 2, 2012 (6-3)
Goal:
Ligate the digested plasmid with the PCR product.
We followed the same protocol listed here. BR
Transformation of Ligated Plasmid into DH5α E. coli cells - May 4, 2012 (6-4)
Goal:
Get the E. coli to uptake our plasmid.
We followed the same protocol listed here.
Transformation with ligated plasmid (6-4A)
Transformation with control (plasmid with no insert) (6-4B) BR
PCR of Colonies (Unsuccessful) - May 7, 2012 (6-5)
Goal:
PCR the DNA of the transformed colonies.
We followed the same protocol listed here. BR
Ran gel of Colonies PCR - May 8, 2012 (7-1)
Goal:
To compare bands and find which colonies have the appropriate DNA band.
Results:
There are too many bands, so our PCR did not work. After the ladder in the first column, we have 8 of our colonies and then our control (malaria group occupies lanes 2-10). Dr. Grose said she would help us do it again, so she can see what may be going wrong. BR
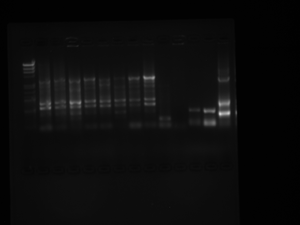
QBP PCR - May 8, 2012 (7-2)
Goal:
Run a successful PCR of QBP.
We followed the same protocol listed here. BR
Purifying Our PCR Product (QBP forward & reverse)- May 9, 2012 (7-3)
Goal:
Successfully purify our PCR product.
We followed the same protocol listed here. BR
We prepared an agarose gel to run out our purified PCR product (7-3).
No band was present on gel. JA
PCR of QBP- May 14, 2012 (8-1)
Goal:
Successfully purify our PCR product.
8-1A= boiled DH5α E. coli template
8-1B= PCR product
We followed the same protocol listed here. BR
After running our PCR product out on gel, we had a band around 700 base pairs, which is the correct length. JA
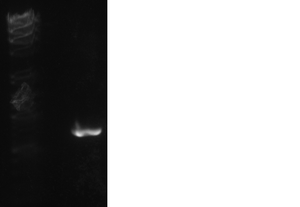
Purifying Our PCR Product - May 15-18, 2012 (9-1)
Goal:
Successfully purify our PCR product.
We followed the same protocol listed here. BR
We prepared an agarose gel to run out our purified PCR product. BR
May 18, 2012 9-1 was left out over night, so we are running it out on a gel again to confirm presence of QBP. There was a band in the correct place (around 700 base pairs), so the PCR product is still good. Just in case the left-out PCR product was not good, we had already started another PCR (9-2). BR
PCR of QBP- May 18, 2012 (9-2)
Goal:
Run a successful PCR of QBP.
We followed the same protocol listed here. We used template 8-1A. JA
PCR Purification of QBP (9-1)- May 22-29, 2012 (9-3)
Goal:
Cleanup and Purify PCR reaction (9-1).
We followed the same protocol listed here. In previous PCR purifications, we would add loading dye to the entire sample after purifying it, but we should only have added it to the small sample that we were going to run out on gel. We re-purified our sample to get rid of the loading dye. BR
May 23, 2012

We ran an agarose gel with the ladder (lane 1), 9-3 (lane 2), control of 9-2 (lane 3), and 9-2 (lane 4). 9-3 had a band in the correct place, but we think we might have mixed up lane 3 and lane 4, so we need to run the control of 9-2 and 9-2 again on gel.
May 29, 2012
We re-ran the gel of 9-2 and the 9-2 control, but it didn't work, so we need to start another PCR reaction.
Restriction Digest of PCR product and plasmid (10-1) - May 23, 2012
Dr. Grose set up our restriction digests for us. We followed the same protocol found here. BR
restriction digest of QBP (9-3)=10-1
restriction digest of pIG12=pIG12 R.D.
Low-Melt Gel - May 25, 2012 (10-2)
Goal:
To run out our digested PCR Product (10-1) and our digested plasmid (pIG12 R.D.) on a low-melt gel, and then to cut those DNA bands out and store them in separate tubes.
Results:
There was only a band for the digested plasmid pIG12 (stored in tube 10-2). BR
PCR of QBP - May 29, 2012 (11-1)
We followed the same protocol listed here. We used template 8-1A. BR
We ran an agarose gel and a band appeared around 700 base pairs, which is the correct length of QBP. BR

Purifying Our PCR Product - June 1, 2012 (12-1)
We followed the same protocol listed here.
Results:
NEEDS PICTURE OF GEL After completing the above protocol we ran the product out on agarose gel and had a correct band (around 700 base pairs). BR JA
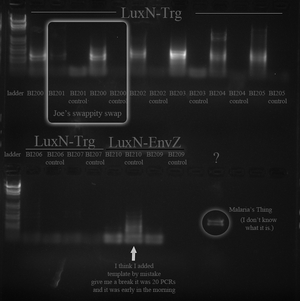
Restriction Digest of Purified PCR Product (12-1) - June 4, 2012 (12-4)
We followed the same protocol listed here. Jordan helped us with the restriction digest to make sure we weren't doing something wrong. BR
Low-Melt Gel - June 5, 2012 (13-4)
Goal:
To run out our digested PCR Product (12-1) on a low-melt gel, and then to cut the DNA band out and store it in a tube.
Results:
Successfully ran the gel and got a band of the expected length (700 bp). We cut out the band and stored it in tube 13-4. BR
Ligation of digested Plasmid and digested PCR product - June 5, 2012 (13-5)
Goal:
Ligate the digested plasmid with the PCR product.
Reagents and Protocol:
1. Melt Gel at 65°C (QBP 10-2, pIG12 13-4)
- Water (6.5μL)
- 10x Ligase Buffer with ATP (1.5μL)
- T4 DNA Ligase (1.0μL)
- digested pIG12 (13-4) (3μL)
- insert (digested QBP 10-2) (3.0μL)
(Control does not include the above insert)
2. Incubate at room temperature for at least 30 minutes. BR
Transformation of Ligated Plasmid into DH5α E. coli cells - June 5, 2012 (15-1)
We followed the same protocol listed here. BR
PCR of Colonies - June 6, 2012 (15-2)
We followed the same protocol listed here. We were surprised to find that some E. coli grew up on our control plate. We took one of the colonies from our control plate for our control in the colony PCR. BR
QBP overnight - June 11, 2012 (15-5)
Goal:
Set up an overnight of colonies 3 and 4 to verify that they are good.
Results:
We could not find the plate that we needed with our colonies so we re-plated the 8 colonies that were marked on the previous plate (15-1). This plate is in the 37 degree on the Igem shelf (labeled: QBP 6/11/12). We used the remaining bacteria from colonies 3 and 4 in overnight tubes. (15-5). JM JA
Plasmid Prep - June 12 and 18, 2012 (15-6)
We followed the protocol listed in the Sigma-Aldrich mini prep kit to prep our plasmids with the QBP insert. We used the overnights from colonies 3 and 4 for the plasmid prep. BR JA
June 18, 2012
We set up the sequencing reactions (labeled 15-6 (3) for colony 3 and 15-6 (4) for colony 4).
Sequencing Reaction Protocol:
- ddH2O (3 μL)
- template (2 μL)- either tube "3" or "4"
- IG57 (1 μL)
BR JM
Title - date (page number)
Goal:
Write goal here.
Results:
write results here (tube number here).