User:Yeem/20.181/10-25
From OpenWetWare
Jump to navigationJump to search
Dihedrals
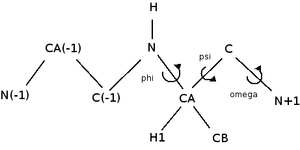
Where CA is the alpha carbon, C(-1) is the C before the current carbon, etc.
Structure of data files for protein backbone:
| Angle | First atom | Second | Third | Fourth |
|---|---|---|---|---|
| [math]\displaystyle{ \phi }[/math] | C-1 | N | CA | C |
| [math]\displaystyle{ \psi }[/math] | N | CA | C | N+1 |
| [math]\displaystyle{ \omega }[/math] | CA | C | N+1 | CA+1 |
Now we want to add other stuff.
| Atom | First atom | Second | Third | Fourth | Dihedral angle |
|---|---|---|---|---|---|
| Amide hydrgeon (NH) | CA-1 | C | N | NH | dihNH |
| Alpha carbon hydrogen (H1) | C-1 | N | CA | H1 | dihH1 |
| First carbon on R-group (CB) | C-1 | N | CA | CB | dihCB |
Cool. But do we always need to calculate stuff from the inputs? Not for the so-called "improper dihedrals", such as the C-CA-N-NH dihedral, or the C-CA-N-NH dihedral, because these are always the same. The "proper" dihedrals (N, CA, C) must be calculated from inputs, but the "improper" dihedrals can be taken from a table.
Build order
N terminus to C terminus, with increasing i
- Within the residue, calculate proper dihedrals, take improper dihedrals from a table