User:Student 63/Notebook/Biology 210 at AU
Food Web
3 March 2016
purpose The purpose of creating a web is to track and visualize the relationships of the biotic factors in our transect
Materials and Methods
Used the data sets from the previous expirements (I-V) and decide the relationship between the organisms within our Transect
Results
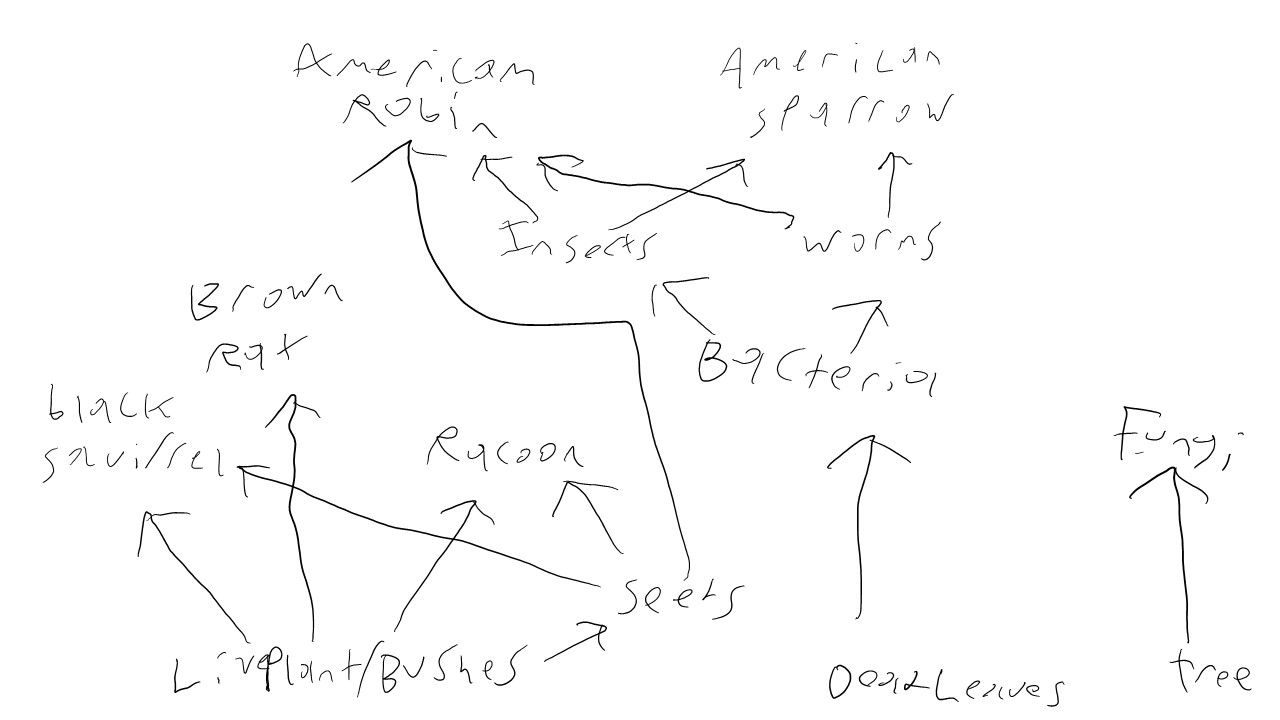 Purpose
Looking at the food web for transect 1 helps us to understand the relationship between the organims living there, and understand how more complex life can live on the foundations of other organisms
Purpose
Looking at the food web for transect 1 helps us to understand the relationship between the organims living there, and understand how more complex life can live on the foundations of other organisms
Max LEopold
16S rRNA gene Results
25 February 2016
Purpose The purpose of this experiment was to analyze the new data found when putting the bacteria from our transect through PCR
Materials and Methods
After watching our bacteria colony grow, we took samples from both of our colonies and inserted them into a gel sample from a Polymerase Chain Reaction. From these, we would hope to observe the expression sequences of specific genes within the bacteria.
Altough our PCR did not work, a fellow lab member's PCR is featured in the text below:
Observations
Unfortunately our transect did not produce bacteria sufficient enough to create a PCR, but looking at Eric Mconough's transect, we have found a similiar reading from a transect of a similar biome that reads as flows:
"Samples 1 and 4, which were run on agarose electrophoresis gel last week and then sent out for sequencing, returned the following sequences, in which N represents unidentified base pairs. Sample 1: TGNANGCCNANCGNGTNAGANGANCGNNNTNCTGNGGNANNCTNTGNGNNAGCGNGNTGATACGGGTGCGGAACACGTGTGCAA CCTGCCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATATTGAATGGCATCATTTGATATTGAAAACTCCGGTGGATA GAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCAGTGATCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTG GTACTGAGACACGGACCAGACTCCTACGGGAGGCAGCAGTGAGGAATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGAC GACGGCCCTATGGGTTGTAAACTTCTTTTGTATAGGGATAAACCTTTCCACGTGTGGAAAGCTGAAGGTACTATACGAATAAGCACCGGCTAACT CCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGATCTGTAAGTCAGTGGTGA AATCTCATAGCTTAACTATGAAACTGCCATTGATACTGCAGGTCTTGAGTAAAGTANAAGTGGCTGGAATAANTAGTGTANCGGTGAAATGCATAG ATATTACTTANNAACACCAATTGCGAAGGGCAGGTCNCTATGTTTTAACTGACGCTGATGGACGAAAGCGTGGGGAGCGAACAGGATTANATACC CTGGTNGTNNNNGCCGTANACGATGCTNACTCNTTTTTNGNNCTTCNGATTCAGAGACTAAGCNAAANTGATAGTTAGNCNNCCTGGNGAGTNC NTTCNCAANAATGAAACTCANAAGAANTGACGGGGGNCCCNCNCANCCGTGNATTATGTNGTTTAATTCANNNNNCNNNNGNANCCTTNNCNAC GCTTAANNGGGATTGNGGGGGNTTAGANNNNANNNGTCTCNNCATTTCNANNTTCTNCNNGGGNNGNCGGNGGNTGGTCCCCCNNTGTANGNN NNGGTCAAGNACNNGNNGNNCCCNNT Sample 4: GCAGTCNAGCGGATGANANNAGCTTGCTCTTCGATTCAGCGGCGGANGGGTGAGTAATGCCTAGGAATCTGCCTATTAGTGGGGG ACAACGTTTCGAAAGGAACGCTAATACCGCATACGTCCTACGGGAGAAAGCAGGGGACCTTCGGGCCTTGCGCTAATAGATGAGCCTAGGTCG GATTAGCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCCGTAACTGGTCTGAGAGGATGATCAGTCACACTGGAACTGAGACACGGTC CAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGATT GTAAAGCACTTTAAGTTGGGAGGAAGGGTTGTAGATTAATACTCTGCAATTTTGACGTTACCGACAGAATAANCACCGGCTAACTCTGTGCCAN CAGCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTCNTTAAGTTNNATGTGAAATCCCC NGGCTCNACCTGNNAGCTNCNTTCNANACTGTCNNGCTANANTATGGTANANGGTGCTGGAATTTCCTCTGTNGNNNNNCAAATNNNNANAT ANANNATTGAACNNNNNTGNCNANNNGNNNNCCTCCNCTGNTNGNCNANNCNNGTATNCGCNCCNTGNGNGNGAAACACNNGNGGNTNGN CTCCNNCGCCACNGNGTNTNCNANATNTACNANCCTTTTCGNTGNGNTNNNNNNNTATTNCNCNGNCTCNCCNNANANNTNNACNNNCANN NTNNNGNNNNGNCCNCCCTGGGGGCTCNNCNNGNNTTTTTNNNTGNCNNGNGCAAGNAANNNNNGNGGGNGTTGNNGCTNNNTNNNACAA AANNANNNNANCGCCCCNTGGTNNNGNNCNGGNNNANGGNNNNTAANANNTGGNNTGGNNNCCTNCGGGNACCNAAANNNNNGGNNGNA NNGNANNNCNNNNNCTCCCNNNCCNANNNNGGTTGG Sample 1 was considerably more definitive than Sample 4, which ultimately could only be fit to a best match of 59% due to the large number of unidentified base pairs. The table below presents the findings when each sample was run through the GeneWiz and Genomic Blast programs. Sample # Number of base pairs Identity of Bacteria % match? Sample 2 1386 Chryseobacterium sp. WR1 80% Sample 3 1397 Pseudomonas sp. Ata11 59% Sample 1 was identified as Chryseobacterium with an 80% match to the known genetic code. Chryseobacterium is gram-negative, rod-shaped, and yellow-pigmented. It is found in environments much like that of this transect, including in soil, plants, and wastewater. This sample had been characterized under the microscope as coccus-shaped, gram positive, and orange-yellow in color. As the color matches, and sample is matched relatively well to the known genetic code, it is likely that this sample is indeed Chryseobacterium, and that some error was involved both in observing the shape of the bacteria under the microscope (identification is difficult for such small organisms without use of an electron microscope) and in the gram stain procedure. Sample 4 was identified with less certainty (59%) as Pseudomonas. Pseudomonas are gram-negative and rod-shaped. As this sample was characterized under the microscope as rod-shaped but gram-positive, and the percent match to the known genetic code is so low, it is likely that this is not a correct identification of this bacteria."
Conclusion Since our PCR did not give us a desired result, our team was unable to make any concrete conclusions from our transects bacteria.
Max Leopold
Exercise V: Invertebrates and Vertebrates
11 February 2016
Purpose
The purpose of this experiment was to characterize the different animals, both vertebrate and invertebrate, within our transect.
In doing so, we learned about the different germ tissues, different mechanisms of mobility, and learned how to recognize the symmetry of animals.
Materials and Methods
Last week we set aside a group of leaves to be dried out over the course of the week, with the results being that animals would fall from the dried leaves into a flask of water sitting below. This week we took the flask of water, separated the sections, top, middle, and bottoms, and observed the biological factors under a microscope to see the different invertebrates that were living within our transect. After making observations, we classified the animials we saw using a provided list in lab.
Observations
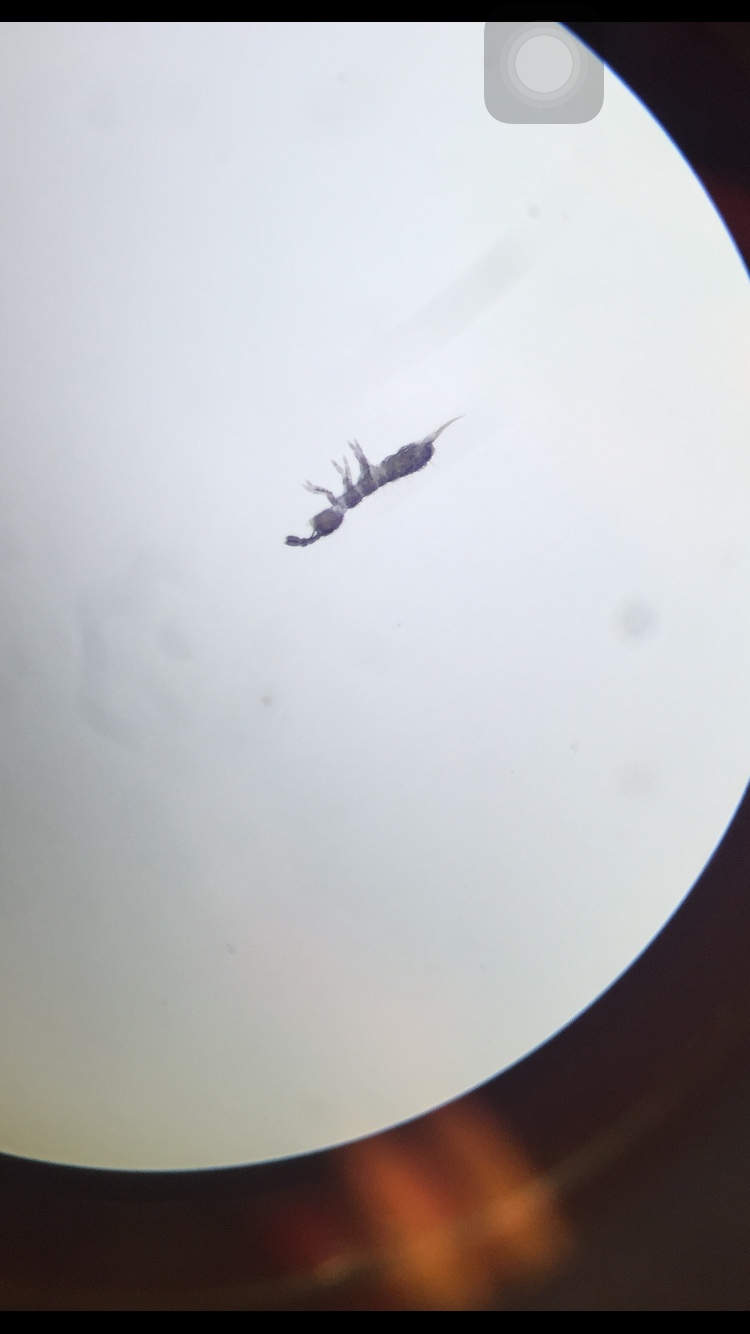
1. 2 Springtails, 2mm, 6legs, wings by its side, light brown/beige, and bilateral
2. 1 Biting lice,.5mm, 6 legs, Orange, bilateral
3. 4 Ticks,.2mm, small, hard shell, 6 legs, no wings, black
4. 1 Termite, .5mm, white
5. 2 Ants, 1.5mm, 6 legs, bead-like antennae, noticeable eyes, brown
Conclusion
Due to the amount of animals found within our transect, and the amount of each animal observed, we were able to conclude that this meant our transect was vary diverse. This may be that previous observations on different plants and antibiotic factors provided a environment for a plethora of different animals to live in.
Max Leopold
Exercise IV: Plant and Fungi
4 February 2016
Purpose
The purpose of this experiment was to characterize the plants and fungi within our transect.
We did this by giving unique examples of plants that had evolved through evolution, decried the differing characteristics between fungi and plants, and distinguished between angiosperm and bryophytes, giving specific examples of each. We also drew a schematic of the alteration of generations in bryophytes or angiosperms, and identified the differing parts of a flower.
Materials and Methods We located and collected 5 plants within our transect, identifying the major groups and genus for each plant, as well as other useful data and descriptions about the plants. In doing so, we described the plants vascularizations, mechanism of reproduction, and any specialized structures. We also noted where they were acquired from within the transect. Lastly, examined the leaves of the bottoms of the transect, and set those up for the next experiment to examine invertebrate.
Observations
Plant #1: Found on the ground near the sidewalk. Decription: long, dark green, parallel lines. Length: 0.5cm wide and 14.1cm long. Monocot and its vascularization was plant.
Plant #2: Found near the back under a bush. Description: light green with intricate lines, ruffled and spiny edges Length: 4.5cm long and 1.3cm wide. It had stomata Dicot and the vascularization was vascular bundles.
Plant #3: Found on the ground. Description: broad leaves,pointy tip, light green Length: 10mm long and 7mm wide. Dicot and its vascularization was vascular bundles
Plant #4: Found on the far corner of transect (Thought to have come from outside transect). Description: Green, glossy color, a single line through the middle, long and thin shape Length: 1cm long and .1cm wide. Monocot with random vascularization
Plant #5: Found on the ground under the light. Description: Light brown on the inside and a darker brown on the outside Length: 3.8cm long and .5cm wide. Had a xylem and phloem Dicot and ring of vascular bundles
Conclusion
Max Leopold
'28 January 2016
This lab we were tasked with observing Bacteria, a prokaryotes, and one of the three domains of life. Bacteria, when exposed to a surface such as an agar plate, wil from a morphology known as a colony, which is a unique characteristic for the domain that shows that organism's growth. For our experiment, we charted the growth of bacteria from our hay culture of last week, and obsessed how the bacterium grew in conditions with and without tetracycline.
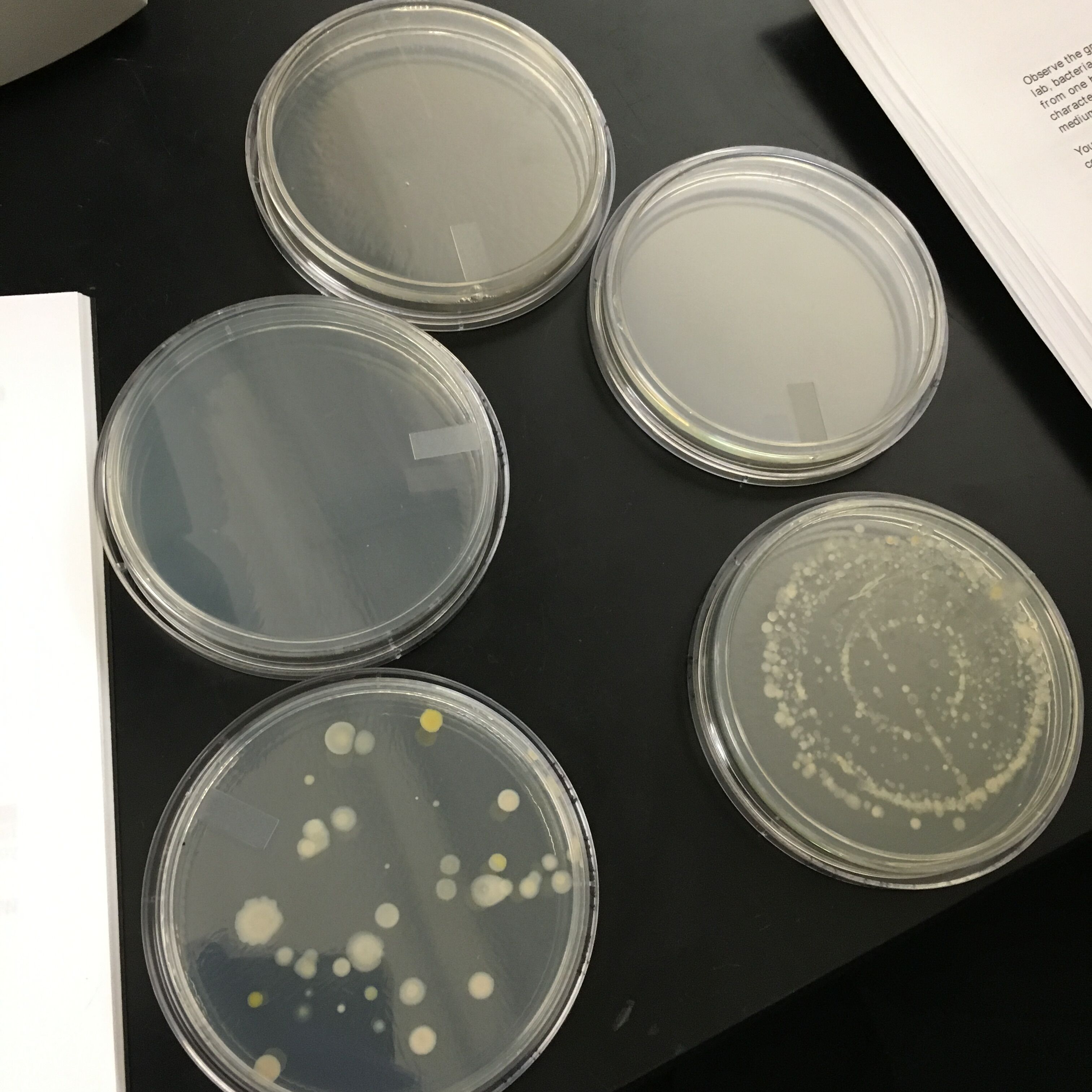
When first seeing the plates that had grown since our previous lab, only two plates, 10^-3 and 10^-5 nutrient only plates, had developed colonies on them. WE first documented how many colonies were on the agar plate at the start of the experiment. After counting we then proceeded to observe the bacteria in oil immersion, observing them at both 40x and 100x focus. Lastly, we finished with a Gran Stain procedure on the bacteria. This involved us coating the bacteria with a color stain that would be used in the Polymerase Chain Reaction (PCR) amplification later. The PCR separate specific genes within the bacteria,, the grain stain dye will help highlight which gense expand when exposed to the PCR, allowing us to identify if the specific bacteria has a specific gene sequence. Specifically, it would be selected to amplify the 16S rRNA gene.
The results of the experiment for the Bacteria were Dilution # of Colononies Colonies per mL 10^-3 880 880,000 10^-5 35 3,500,000
The results from the oil observation showed:
Colony Type Desciption Cell desprition Gram + or -
10^-3 nutrition clear, ciruclar, small circles rod shaped, no movement +/-
10^-5 nutrition clear, circiular, sphere, no movement -
ML
21 January 2016
The hay culture was made using a smaple of the transect described last week, as well as .1 gram of dried mil, and 500 mLs, and was set aside for a week as the cultures developed within the transect. The observations were:
-The smell of the hay culture was a mixture of moldy and mildew. Upon observing it, the hay culture appeared to have a light brownish color, with lots of settlement at the bottom of the jar, and a dark film around the top of the jar.
-There was a bit of greenish mold on the top of the water and on the glass around the rim
-Closer to the plant matter might have more developed cultures thanks to the ability for organisms to develop on a solid surface near nutrition.
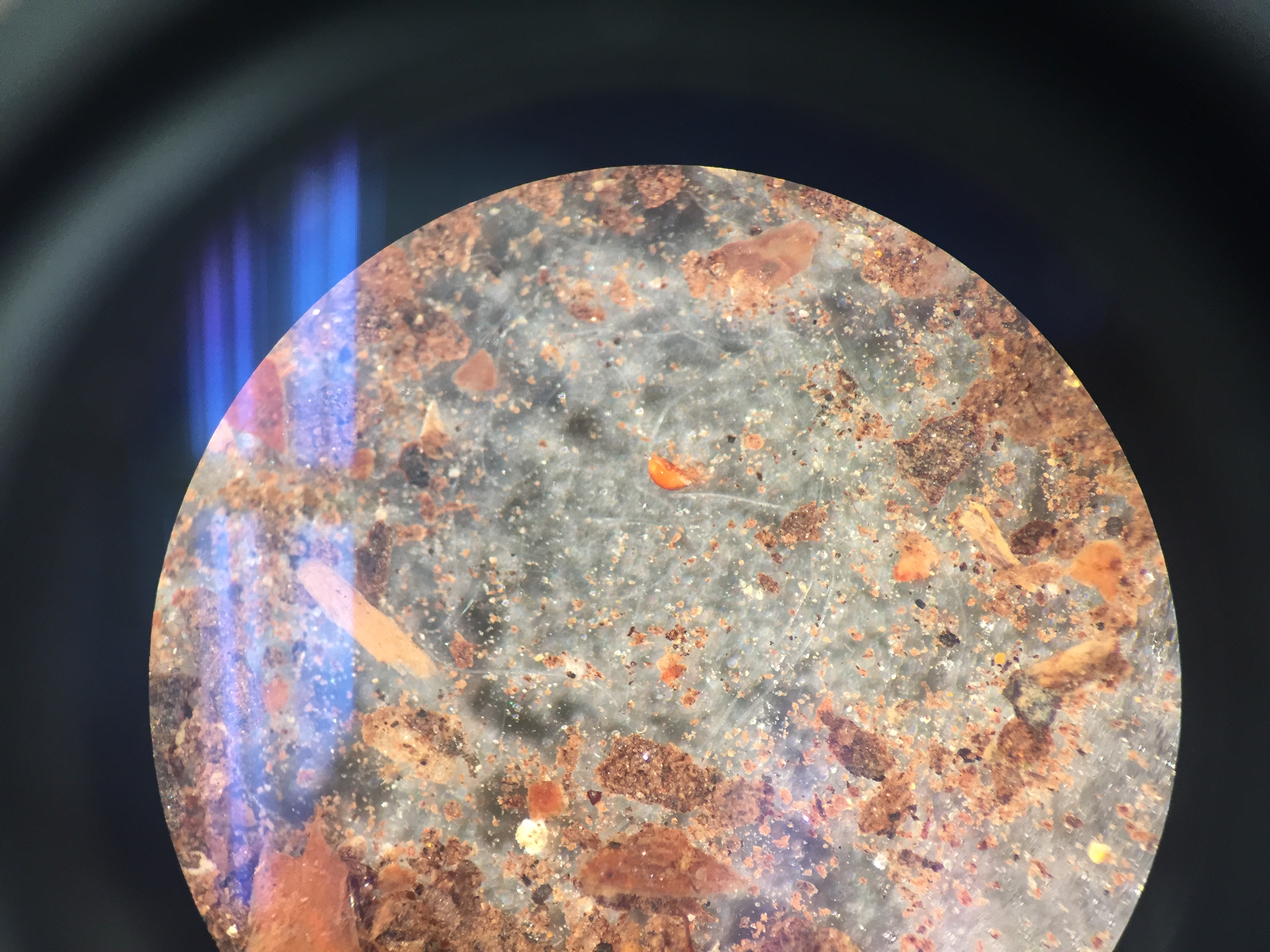
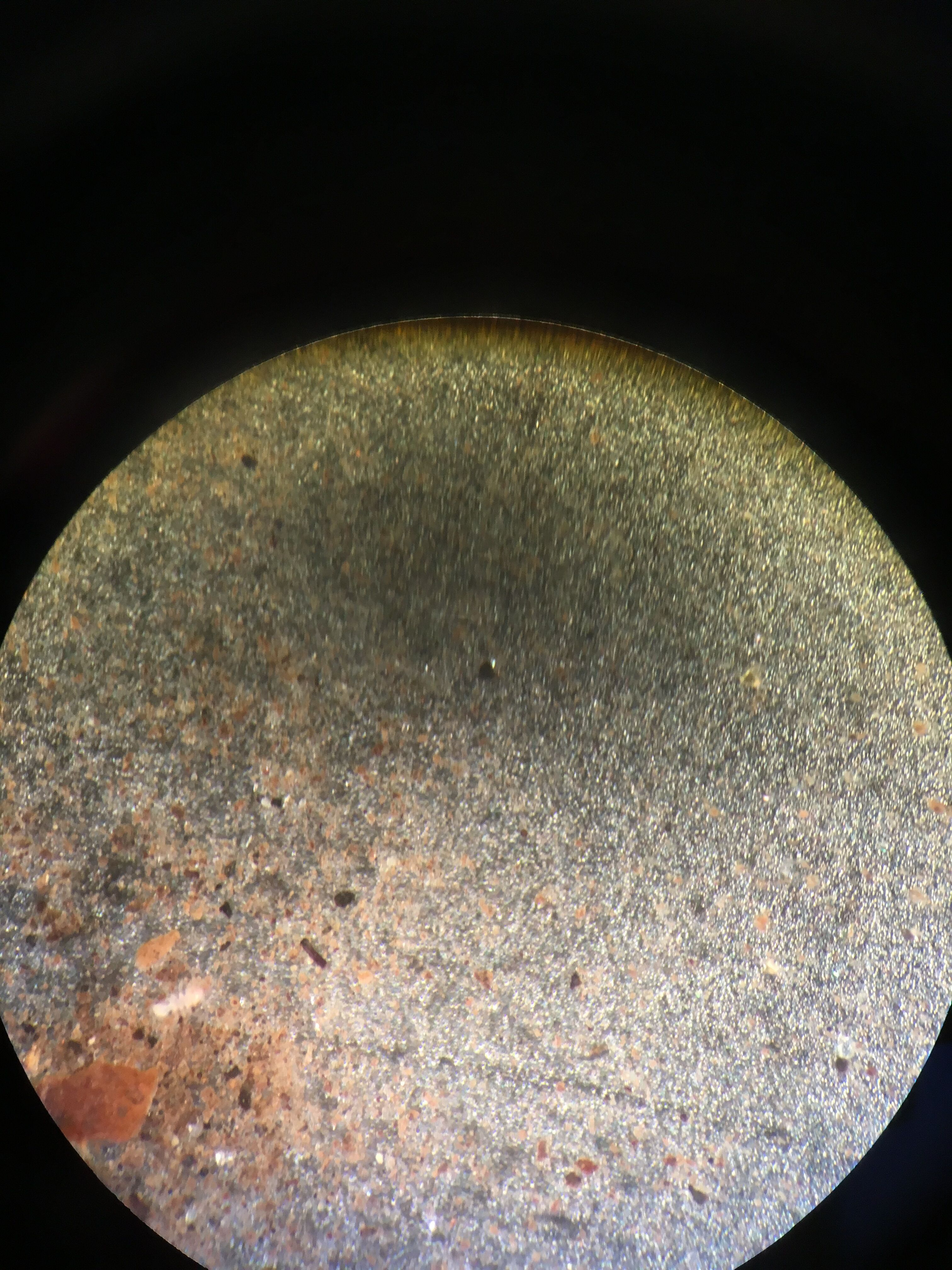
-Our first selection was on the top layer, where we found: Diatoms brownish creatures with small dots and plated groves, measuring 1,500 micrometers [[Image:]] Pelomyxa small, motionless colonies which consistently changed shape, measuring 30 micrometers [[Image:]] Chlomydomonaswhich were small, round, and clear organisms, measuring 25 micrometers [[Image:]]
-Our second selection was the middle layer, where we found: Pandorina measuring between 25 and 40 micrometers [[Image:]] Euglena green, single cellular with one flagella, measuring 10 micrometers [[Image:]] Pandorina colony of small cells in spherical shapes, measuring about 30 micrometers [[Image:]]
-The Chlomydomonas, as described by Freeman, exhibit all the criteria for life. As is clearly observable, the organism is made of a cell, completing that criteria. Finally, it's movement by Flagella also exhibit the use of energy crossing off that part of criteria. The cell also shows evidence that it evolved from some pre existing cell, which shows that it came as a product of evolution as well as capable of replication. When it ovoids the objects around it, the cell also displays it's ability to process information.
-If left unobserved for two months, their would be some significant changes. First, their would perhaps be more, and possibly more complex, creatures in the hay culture. The abiotic factors such as the first and dead leaves on the bottom of the jar would likely have decomposed and denatured, or changed as the culture in the jar changed. The extend length of time would put a lot more factors into the growth and development of the culture, such as fluctuation in temperature and sun's position causing either difficult or more prosperous growth.
ML
14 Janurary 2015
Transect 1: 20x20 square feet plot of land outside of Hurst, directly next to the ward circle building. The transect is largely covered with dirt and dead leaves, as well as the small shrubbery around the system. The two defining pieces within the transect are the large tree in the middle of the transect, as well as the large lamppost within the area, which provides a constant source of light within the transect. This might detract a lot of large fauna might be deterred from the area. However, this combined with the sprinklers provide a constant source of water and light for the plants in the area. The transect is also next to a paved road, which provides a lot of foot traffic next to the transect, and might contribute a lot of human litter into the transect.
Biotic factors: About 8 small trees (known as “Kerria Japonica ‘Plenifora; from the sign near the transect), Large tree in the center of the transect, cut down shrubs, as well as grass, nuts, and dead leaves on the ground of the transect.
Abiotic factors: Light post (with a constant source of light), sprinklers (with a constant source of water), Litter (including cigarettes and straws), and a large cobblestone wall in the back
ML