User:Pedrobeltrao/Notebook/Structural analysis of phosphorylation sites/domain based analysis
Domain based analysis
Phosphorylation sites from S. cerevisiae were collected from different sources . In addition phosphorylation sites from H. sapiens, D. melanogaster, C. albicans and S. pombe were transfered by homology to S. cerevisiae proteins by aligning S. cerevisiae proteins with orthologous proteins in these species. Up to date values for phospho-sites can be seen [here]).
One possible type of analysis that can be done with this information is to predict modes of regulation for protein domains that are commonly phosphorylated. For each PFAM domain we can we have picked an example structure to use as a structural model. We then mapped all phosphorylation sites that occur in instances of these domain in S. cerevisiae or orthologous proteins to this model structure and calculated phosphorylation propensity . As an example we show below the results obtained for the protein kinase domain.
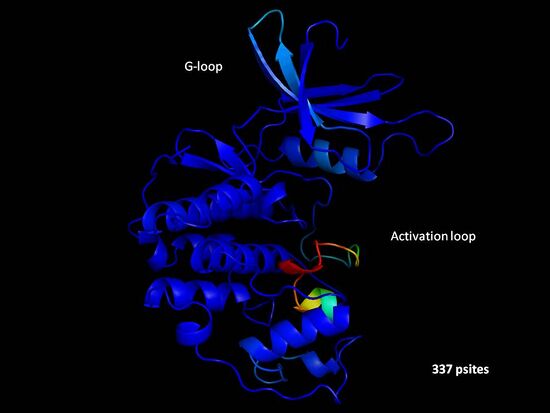
In this example the phosphorylation propensity matches well with the known modes of regulation of protein kinases. The phosphorylation of the activation loop (in red) with the highest propensity as well as a moderate phosphorylation propensity of the glicine rich loop (light blue). We are currently analyzing other domains for to search for novel regulatory sites.