User:Natalia Pabon-Mora
Contact Info

GRUPO EVO-DEVO EN PLANTAS, UNIVERSIDAD DE ANTIOQUIA (IP:Natalia Pabon-Mora)
- Address:
Instituto de Biología Universidad de Antioquia Medellín, Colombia AA 1226
The Plant Evo-Devo Research Group at the Universidad de Antioquia (UdeA) started in 2012. Our group is interested in studying the comparative morphology and genetic basis of leaf, flower and fruit development of non-model, Neotropical plants. We are interested in exploring the effect of gene and genome evolution in plant morphology and diversification. Many of our projects aim to assess how genes and protein networks responsible for floral organ identity and fruit development, that for the most part have been identified in model species (like Arabidopsis thaliana) have changed during plant evolution. We study naturally occuring mutants, ontogenetic shifts that may have enhanced speciation and natural shape and structural variation in flowers and fruits of taxa that share a common ancestor.
Our projects are done in collaboration with Dr. Favio González (Universidad Nacional de Colombia), Dr. Juan Fernando Alzate (Centro Nacional de Secuenciación Genómica), Dr. Lucia Atehortúa (Biotechnology Research Group UdeA). Collaborators abroad include Dr. Sabine Zachgo (University of Osnabrueck), Dr. Barbara Ambrose (The New York Botanical Garden) and Dr. Amy Litt (University of California, Riverside). We also collaborate closely with Dr. Dennis Stevenson (The New York Botanical Garden) and Dr Gane K-S Wong (University of Alberta) and use the resources made available by the oneKP project (https://sites.google.com/a/ualberta.ca/onekp/).
Education
- 2012, PhD, The City University of New York (CUNY) and the New York Botanical Garden (NYBG)
- 2009, MS, City University of New York (CUNY)
- 2006, BS, Universidad Nacional de Colombia
Research interests
Funded projects are related with the following topics:
*1 The morfo-anatomical and genetic basis of organ identity and functional transfer in floral whorls in the Aristolochiaceae.
The ABCE model of flower development was postulated based on floral mutants of the core-eudicot Arabidopsis thaliana. The model predicts that floral organ identity in Arabidopsis requires the specific overlapping expression of four classes of MADS-box transcription factors and the occurrence of specific tetrameric protein interactions in each floral whorl. Thus, E class genes are responsible for proper floral meristem and organ identity in all whorls. Coupled with them, A-class genes alone control sepal identity, A+B-class genes control petal identity; B+C-class genes control stamen identity, and C-class genes alone control carpel identity. However, comparative analyses have shown that the model cannot be easily extrapolated to other flowering plants, mostly due to MADS-box lineage duplications followed by divergent functional evolution during angiosperm diversification. We are studying the expression of ABCE-class and AGL6 homologs in the basal angiosperm Aristolochia fimbriata (Aristolochiaceae), an herb with fast life cycle and unique flower morphology among Magnoliids. We are also exploring the interactions between the A, B, C, E and AGL6 floral proteins in yeast and will compare their interaction capabilities with those reported in core-eudicots.
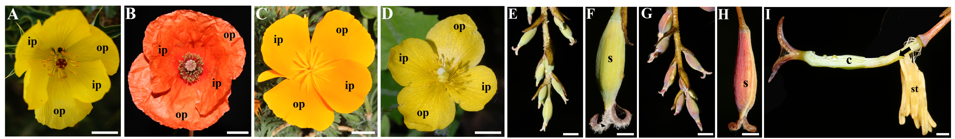
*2 The developmental and genetic explanation of the loss of petals in wild poppies.
With c 760 species, the poppy family (Papaveraceae) exhibits a dimerous floral plan with 2 deciduous sepals, 4 petals in two whorls, many stamens (up to 700) and 2(-8) fused carpels. The New World Bocconia Plum. ex L. (10 spp.) and its sister temperate genus Macleaya R. Br. (2 spp.) are the only genera with apetalous flowers. We are investigating the morphological and genetic bases responsible for the apetaly in Bocconia frutescens L., which are likely the same in Macleaya cordata R. Br.. Homeosis and gene loss of some members of the B-class candidate genes likely account for petal loss.
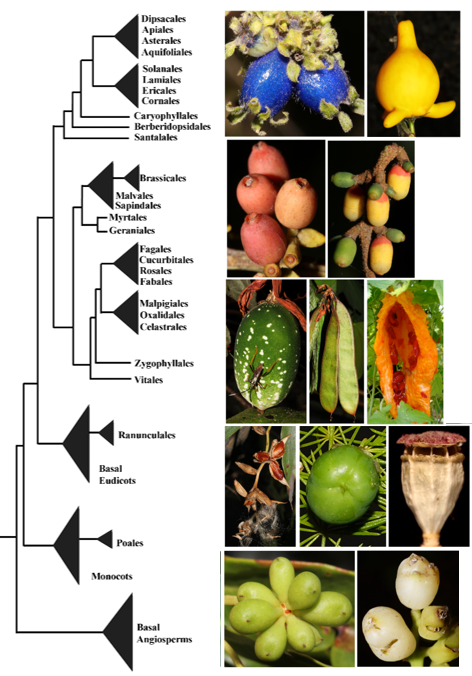
*3 The evolution of the genetic regulatory network of fruit development across angiosperms.
Fruits are one of the most incredible examples of extreme differentiation throughout ontogeny,as despite the fact that they all form from the gynoecia (and sometimes accessory parts), they develop into incredibly plastic structures for seed dispersal. The genetic mechanisms regulating dry fruit development and opercular dehiscence have been identified in Arabidopsis thaliana, and functional evidence suggests that they are fairly conserved across Brassicaceae. In the bicarpellate silique, valve elongation and differentiation is controlled by FRUITFULL (FUL), that antagonizes SHATTERPROOF1-2 (SHP1/ SHP2) and INDEHISCENT (IND) at the dehiscence zone, where they control normal lignification. SHP1/2 are also repressed by REPLUMLESS (RPL), responsible for replum formation. Similarly, FUL indirectly controls two other factors ALCATRAZ (ALC) and SPATULA (SPT) that function in the proper formation of the separation layer. Some of these transcription factors are known to be the result of Brassicaceae specific duplications, others seem to be the result of duplications coinciding with the origin of the core eudicots. We are studying the evolution of APETALA2 FRUITFULL, SHATTERPROOF, ALCATRAZ, SPATULA and REPLUMLESS to better hypothesize what parts of the network for fruit development in Brassicaceae, might be conserved across Angiosperms.
*4 The extreme morphological changes in inflorescence and flower construction in Neotropical parasitic plants.
Santalales include c.a. 2200 species in 18 families, 11 of which are Neotropical. The habit, inflorescence and flower morphology, embryology and pollen, provide distinguishing characters among families and genera. Current phylogenetic analyses have reorganized the interfamilial relationships, making necessary a reassessment of the ontogeny, morphology and character evolution especially in Loranthaceae and Viscaceae. In particular, character evolution associated with the inflorescence architecture, floral sex, and stamen morpho-anatomy needs to be tested. We are studying the development of inflorescences and flowers in Gaiadendron punctatum G. Don., Phthirusa stelis (L.) Kuijt, P. pyrifolia Eichler., Aetanthus mutisii Engl., Oryctanthus sp. (Loranthaceae), and Dendrophthora avenia (Trel.) Kuijt. and Phoradendron nervosum Oliv. (Viscaceae). These species are stem hemiparasites except G. punctatum a root hemiparasite. They have in common the indeterminate vegetative growth, epipetalous stamens, superior gynoecium and berries. We are finding a number of unique ontogenetic features in Viscaceae that may help explaining the modular inflorescences and the floral reduction typical in the family.
In addition we are studying the occurence of Pilostyles (Apodanthaceae) and Castilleja (Orobanchaceae) in dry forests and paramos, and we are beginning to explore their parasitic associations with Dalea (Fabaceae) and Plantago (Plantaginaceae) respectively.
Publications
Horn, S., N. Pabón-Mora, V. Theuß, A. Busch, S. Zachgo. Analysis of the ancestral CYC/TB1 class TCP transcription factors in basal angiosperms and magnoliids. The Plant Journal. In press.
Pabón-Mora N., G-K S Wong, B.A. Ambrose. 2014. Evolution of fruit development genes in flowering plants. Frontiers in Plant Science. doi:10.3389/fpls.201400300
González, F., Callejas-Posada R., N. Pabón-Mora. 2014. Rediscovery and conservation status of the “cloud fern,” Nephopteris maxonii (Pteridaceae), with notes on its anatomical traits. Brittonia. In press.
González,F., N. Pabón-Mora. 2014. First reports and generic descriptions of the achlorophyllous holoparasites Apodanthaceae (Cucurbitales) of Colombia. Revista Actualidades Biológicas 36: 123-135.
González, F., N. Pabón-Mora. 2014. Pilostyles boyacensis, a new species of Apodanthaceae (Cucurbitales) from Colombia. Phytotaxa 178 (2):138-145
González, F, N. Pabón-Mora. 2013. A new species of Castilleja (Orobanchaceae) from the páramos of the Colombian Eastern Cordillera, with comments on its association with Plantago rigida (Plantaginaceae). Caldasia 35:261-272
Pabón-Mora, N., O. Hidalgo, S. Gleissberg, A. Litt. 2013. Assessing duplication and loss of APETALA1/FRUITFULL homologs in Ranunculales. Frontiers in Plant Science. doi: 10.3389/fpls.2013.00358
Pabón-Mora N., S. Bharti, E. Kramer, A. Litt. 2013. The Aquilegia FRUITFULL-like genes play key roles in leaf morphogenesis and inflorescence development. The Plant Journal 74: 197-212.
Pabón-Mora, N., F. González. 2012. Leaf development and metamorphic heteroblasty in Berberis s.l. (Berberidaceae). Botanical Review 78: 463-489
Pabón-Mora, N., B. Ambrose and A. Litt. 2012. Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity and fruit development. Plant Physiology 158: 1685-1704. ISSN: print: 0032-0889, online: 1532-2548.
M. Graça Sajo, N. Pabón-Mora, J. Jardim, D. W. Stevenson and P. J. Rudall. 2012. Homologies of the flower and inflorescence in the early divergent grass Anomochloa (Poaceae). American Journal of Botany 99: 614-628. ISSN: print: 0002-9122, online: 1537-2197.
G. Doria, N. Pabón-Mora, F González. 2012. Reassessing inflorescence and floral morphology and development in Hedyosmum (Chloranthaceae). International Journal of Plant Sciences 173: 735-750. ISSN: print: 10585893 ; online: 15375315.
Pabón-Mora, N. and A. Litt. 2011. Comparative anatomical and developmental analysis of dry and fleshy fruits of Solanaceae. American Journal of Botany 98 (9): 1415-1436. ISSN: print 0002-9122, online 1537-2197.
González, F., H. E. Esquivel, G. A. Murcia & N. Pabón-Mora. 2010. Aristolochia pentandra (Aristolochiaceae) in Colombia: Biogeographic implications and proposed synapomorphies between the pentandrous species of Aristolochia and its South American sister group. Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales 34 (133): 467-478. ISSN: 0370-3908.
González, F. & N. Pabón-Mora. 2009. De Henslow a Hooker: Darwin y los inicios del pensamiento evolutivo en Botánica. Acta Biológica Colombiana 14 (S): 311-334. ISSN: print: 0120-548X online:1900-1649.
Pabón-Mora, N. & F. González. 2008. Floral ontogeny of Telipogon spp. (Orchidaceae) and insights on the perianth symmetry in the family. International Journal of Plant Sciences 169: 1159-1173. ISSN: print:10585893; online: 15375315.
Mora-Osejo, L. E., N. Pabón-Mora., F. González. 2011. Gunneraceae. Flora Neotropica Monograph No. 109. The New York Botanical Garden Press, Bronx, NY, 166 pp. ISBN-10:0-89327-508-5 ISBN-13: 978-0893275082
Pabón-Mora, N., F. González. 2011. A classificação biológica: de espécies a genes. In P. Abrantes (ed). Filosofia da Biologia. pp. 123-144. Universidade de Brasília, Brasil. ISBN: 978-85-363-2451-7
Teaching
UNDERGRADUATE COURSES:
- General Biology. Module: evolution and development, Universidad de Antioquia (1 sem)
- Developmental Biology, Universidad de Antioquia (6 sem)
- Introduction to Biotechnology, Universidad de Antioquia (8 sem)
- Seminar in Plant Developmental Biology, Universidad de Antioquia (prof)
GRADUATE COURSES
- Evolution and Development of tropical plants, Universidad de Antioquia
Group Members
GRADUATE STUDENTS
* Harold Suárez Baron - Early evolution of the floral developmental gene toolkit: a case study in the Magnoliid Aristolochia fimbriata (Aristolochiaceae)
My main interest is to understand the evolutionary dynamics and molecular mechanisms of plant development with a special focus on the floral organ identity in early diverging angiosperms, using a functional genomics and developmental genetics approach. My Master´s thesis project is focused on the study of the expression of ABC and E floral development genes and their interactions in the floral groundplan of Aristolochia fimbriata.
UNDERGRADUATE STUDENTS
*Cristina Arango Ocampo - The genetic and morpho-anatomical basis of the floral homeosis in Bocconia frutescens and Macleaya cordata (Papaveraceae)
*Cecilia Zumajo Cardona - Evolution of the ERF/AP2 gene lineage in seed plants
I am interested in studying the AP2/ERF gene superfamily, a lineage that has undergone numerous duplications, presumably in seed plants. The best known gene in this group is APETALA2, responsible for sepal and petal identity as well as fruit development in Arabidopsis. I am working on increasing the sampling of euAP2 genes across different lineages of seed plants, to develop a more robust phylogenetic hypothesis. In addition I am evaluating the expression of the euAP2 orthologs, in representative taxa of the diversity of flowering plants, to develop a hypothesis of the change of function of these genes during the diversification of seed plants.
*Vanessa Suaza Gaviria - Plant architecture, inflorescence and flower development in Santalales
*Clara Ines Ortiz - Expression and function of ALCATRAZ in fleshy and dry fruits in Solanaceae
*Yesenia Madrigal Bedoya - APETALA3 and TCP-like gene evolution in the Asparagales: exploring a putative link with Orchid diversification
I am interested in the evolution of floral development genes in the order Asparagales. I am studying the evolution of B-class, C-class and CYC/TCP genes across the Asparagales, with a particular sampling in Orchidaceae to test whether these gene lineages have undergone specific duplications in orchids. Additionally I am interested in conducting expression studies of B, C, and CYC/TCP1 genes in neotropical orchid species (modified trimerous flowers) in comparison with members of the Hypoxidaceae (canonical trimerous flowers) to generate hypotheses fucntional changes likely resulting in floral grounplan modifications in the two taxa.
*Pablo Perez Mesa - MADS-box protein-protein interactions in Aristolochia fimbriata
I am interested in the historical processes and the evolutionary patterns that have allowed the diversification of plants. In particular I am interested in the relation between gene evolution and the molecular basis of organ identity in plant development on one side and plant diversification on the other. I am studying the interactions among MADS-box proteins involved in flower development of Aristolochia fimbriata, a basal angiosperm, to test whether the protein interactions at the base of the ACBE model of flower development postulated for model core-eudicot species are maintained in early diverging plant lineages. These data will provide new hypothesis about functional changes and new interactions that MADS box proteins may have acquired (or lost) during the evolution of the Angiosperms, and to test the ABCE model applicability across flowering plants.