User:Matthew R Skorski/Notebook/Biology 210 at AU
3/22/14
Objective: The objective of this experiment was to determine if alcohol has any negative affects on zebrafish development in order for them to serve as a model for fetal alcohol syndrome in humans. Fetal alcohol syndrome is a condition caused by mothers consuming alcohol while pregnant. The detrimental affects of this syndrome are known on humans, such as growth retardation, face abnormalities, sensory deficits, impaired fine motor skills, and learning disabilities. Between ethical concerns regarding giving pregnant mothers alcohol which negatively harm their unborn child and unreliable reports by mothers who did drink while pregnant, a model organism is needed to better understand the effects of alcohol on a growing fetus. Thus, this experiment aims to see if zebrafish can serve as an effective model organism for this condition.
Current literature points to zebrafish being a superior model organism for Fetal Alcohol Syndrome due to it's transparent eggs and ease of introducing the ethanol to the zebrafish. With rat or mice models the pregnant rats must have their blood alcohol checked regularly to see if if the embryos are being exposed to the right concentration of alcohol. Zebrafish by contrast take in the ethanol from their surroundings, preventing the researcher from having to check a mother for blood alcohol levels. Additionally zebrafish can breed large quantities and grow up exceptionally fast, allowing an experiment to be done very quickly and in a large number.
It is known that zebrafish exposed to alcohol while an embryo have higher mortality, smaller eyes, smaller body lengths, changes in heart rate, and more structural abnormalities. The results from this lab will be compared to these results to see if they hold true under additional scrutiny.
Methods
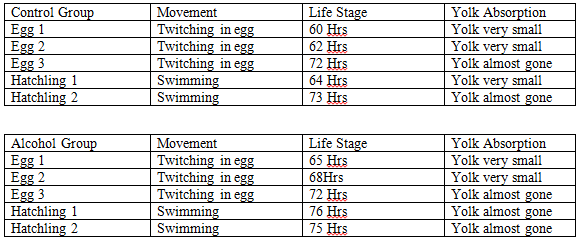
Day 1, 2/19: Two petri dishes were set up, one containing 10 mL of a 1% alcohol solution and a control containing 10 mL of distilled water. The movement and life stage of 5 eggs from each group were analyzed with the naked eye.
Day 3, 2/21: The dead eggs were removed and the number of zebrafish that hatched were counted from each group. The yolk absorption, life stages, and movement of 5 zebrafish were taken with a mixture of egg and hatched fished chosen. They were analyzed with the naked eye.
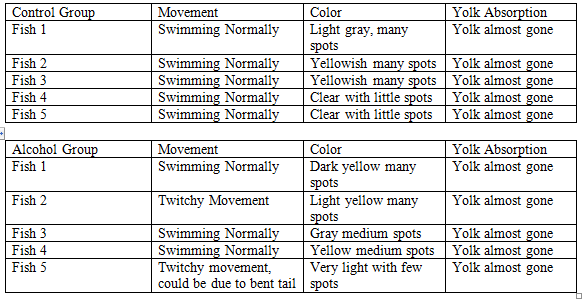
Day 6, 2/24: The solution in each dish was pipetted out and replaced with 10 mL of each respective solution. Any dead fish were removed and disposed of. The number of remaining fish was counted. The movement, yolk absorption, color and pigmentation of 5 zebrafish from each group were analyzed with the naked eye and recorded.
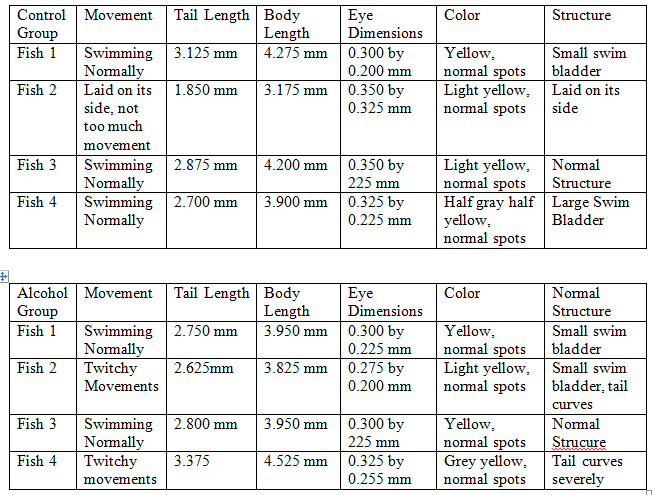
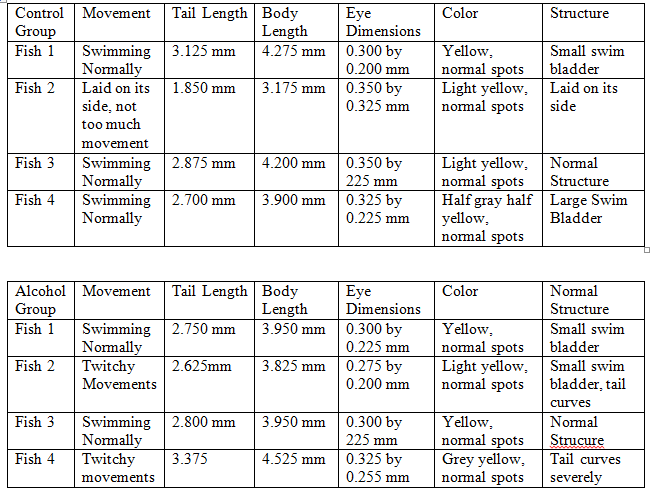
Day 8, 2/26: The number of remaining fish was counted. Four zebrafish from each group were pipetted onto a wet mount and observed under a microscope. The tail length, body length, eye diameter, color, pigmentation, and structure were recorded. Two zebra fish from each observed group were selected to be anesthetized and preserved in formaldehyde.
Day 10, 2/28: The solution in each dish was pipetted out and replaced with 10 mL of each respective solution. Any dead fish were removed and disposed of. Four zebrafish from each group was pipetted onto a wet mount and observed under a microscope. The tail length, body length, eye diameter, color, pigmentation, and structure were recorded. Two zebra fish from each observed group were selected to be anesthetized and preserved in formaldehyde.
Day 14,3/4: Any dead fish were removed and disposed of, with the better preserved ones being preserved in formaldehyde. The solution in each dish was pipetted out and replaced with 10 mL of each respective solution. Any salvageable dead zebrafish were preserved in formaldehyde.
Day 15, 3/5: All the dead fish were disposed of. All the preserved fish from previous experiments were remeasured to insure accuracy.
Results:
Note: Hrs means hours
Day 1, 2/19:
20 eggs are in both dishes.
Day 3, 2/21:
In the control group 1 egg died, leaving 19 zebrafish alive. 10 hatched while 9 remained in their eggs. In the alcohol group 2 eggs died, leaving 18 zebrafish alive. 10 hatched while 8 remained in their eggs.
Day 6, 2/24:
In the control group all the eggs hatched, however 1 fish died leaving 18 left. In the alcohol group all the eggs hatched, however 2 fish disappeared leaving 16 left.
Day 8, 2/26:
In the control group 18 fish were alive, however 1 was lost while analyzing it and 3 were preserved so 14 were left. In the alcohol group 16 were alive, with 3 being preserved 15 were left.
Day 10, 2/28
In the control group 14 fish were alive, two were sacrificed to be preserved so 12 remained at the end. In the alcohol group 2 had died between labs leaving 11 left. Of those 11 two were selected to be sacrificed to be preserved so 9 remained.
Day 14,3/4
There was a mass die off since the last time the zebrafish were observed. The control group had only three zebrafish left and the alcohol group had no zebra fish left. The remaining three zebrafish did not appear to be healthy, they did not move much. They were not measured.
Day 15, 3/5:
The three zebra fish from last time died. Attempts were made to look at preserved samples from Day 14 but the bodies were too decomposed to make accurate measurements.
Overall, the biggest difference observed between the two groups was the movement. The alcohol zebrafish had much twitchier movements than the control. While the control zebrafish would move in clean, fluid motions the alcohol zebrafish would rock back and forth slighlty while moving. From the other variables both groups ran about the same.
Discussion:
As stated previously the most noticeable difference between the two groups was the movement of the fish. The alcohol fish were jerky and would make movements that would remind one of a seizure. The alcohol fish moved slower too, making them much easier to capture than the control fish. While this does not directly connect with what was expected it does represent a negative affect the alcohol group had that the control didn't. Thus, the impaired movement of zebrafish could be factor in fetal alcohol syndrome among zebrafish.
From the data it appears that for tail, body, and eye size the two groups were about even. Interestingly the size of the fish did not appear to go up too much between 2/26 and 2/28. In fact, the average size went down as all the fish from 2/26 had a body length of at least 4.000 mm. The 2/28 fish however, had several fish that did not break that length in both the control and alcohol groups. This could be due to human error as it is unlikely that all the fish shrunk. Instead the fish may have been measured differently between the two days, overestimating on the first and underestimating on the second. Unfortunately, the next day the fish were meant to be observed, 3/4, had a massive die off that killed off the vast majority of the remaining fish. The bodies were so badly decomposed that accurate measurement was impossible. Thus, there is a gap in the data that may have shown results. While the cause of the die off is unknown it obviously was environmental to both dishes as they were almost equally affected. While three control fish did live, as they died by the next day it was determined not to be significant.
Another factor that could have shown a difference was the rate of abnormalities. However, several fish from each group had bent or curved tails as well as abnormalities of the swim bladder with the total being 5 control fish as being not structurally normal and 7 alcohol fish being not structurally normal. Seven to five is not enough data to show that the alcohol made a difference, obviously structural abnormalities are a natural process for it to affect both groups.
The coloring and pigments of the fish appeared normal throughout the experiment with the exception of the control 4 fish from 2/28 that was half gray and half yellow down the middle. While this particular fish did have a large swim bladder it is unknown if the two are connected.
The data on yolk absorption, life stage, hatching, and death rates also appeared to be constant throughout both groups. On 2/21 and 2/23 disappearances and deaths were common in both. No late hatching, yolk absorption, or life stage development was observed among the alcohol group.
While heart rate, eye movement, and eye pigmentation was recommended to be observed all three variables proved to difficult to observe. For many zebrafish the heart was too small to locate and count. Thus, any data that was gathered was made useless as there wasn't much to compare it too. The eyes movement and their pigment suffered a similar issue as the eyes could only be observed from above with no way of knowing if the eyes were different or not besides size. Thus, these three variables were excluded from the data sets.
With all this data analyzed it was soon realized that the main problem of this experiment was scale. The populations of zebrafish used were too small to account for random deaths or mutations that would naturally whittle down the numbers. With only four zebrafish observed critically under the microscope per day it is unknown how the entire population was doing and if the alcohol group did have more abnormalities or defects than the control group. Additionally, with only two days of microscope observation and the massive die off it is unknown if any more symptoms would arise in the alcohol group compared to the control group. It is recommended that this experiment be repeated again, this time with more zebrafish and all the fish observed under the microscope in order to get enough data to see if there are more difference between the the two populations than movement. Increasing the alcohol concentration too could help, perhaps the 1% concentration used was too low. Additionally, the zebrafish should be kept in a more hospitable location in order to prevent another die off which greatly impacted the data collection.
While impaired motor skills is a sign of fetal alcohol syndrome in humans and was observed among the alcohol zebrafish, this set up is clearly not effective enough to serve as a proper model for humans. There were too many errors and not enough evidence such as a size difference or an increase in physical abnormalities to reflect what is known to affect humans. Human error may have been large assessing the size difference as the average size went down and not up as it should have over a two day gap. Nevertheless, if the above measures are followed, zebrafish could be useful. Until then, more research must be done in order to optimize conditions.
MRS
3/7/14
Objective:
The object of this lab was to run the DNA sequence of two samples, the black bacteria colony from 10^(-5) tetracycline free plate and the green colony from 10^(-3)tetracycline plate in order to better determine their species.
Methods:
See Lab 3.
Results:
Green Bacteria Sample:
Sequence: GGTATTAGCTTACTGCCCTTCCTCCCAACTTAAAGTGCTTTACAATCCGAAGACCTTCTTCACACACGCGGCATGGCTGGATCAGGCTTTCGCCCATTGTCCAATATTCCCCACTGCTGCCTCCCGTAGGAGTCTGGACCGTGTCTCAGTTCCAGTGTGACTGATCATCCTCTCAGACCAGTTACGGATCGTCGCCTTGGTGAGCCATTACCTCACCAACTAGCTAATCCGACCTAGGCTCATCTGATAGCGCAAGGCCCGAAGGTCCCCTGCTTTCTCCCGTAGGACGTATGCGGTATTAGCGTTCCTTTCGAAACGTTGTCCCCCACTACCAGGCAGATTCCTAGGTATTACTCACCCGTCCGCCGCTGAATCGAAGAGCAAGCTCTTCTCATCC
Name: Psedudomonas Putida
Qualitative: While this individual strain was not observed under the microscope it was at least determined to be green.
Quantitative: From research, Psedudomonas Putida is a rod-shaped and gram negative soil bacteria.
Because the black bacteria did not give any kind of fragment another group's data from the same transect was used.
Name: S1-T1-12
Sequence: TCAGCTACTCTCACGAGAGTAGGTTTATCCCTATACAAAAGAAGTTTACAACCCATAGGGCCGTCGTCCTTCACGCGGGATGGCTGGATCAGGCTCTCACCCATTGTCCAATATTCCTCACTGCTGCCTCCCGTAGGAGTCTGGTCCGTGTCTCAGTACCAGTGTGGGGGATCACCCTCTCAGGCCCCCTAAAGATCGTAGACTTGGTGAGCCGTTACCTCACCAACTATCTAATCTTGCGCGTGCCCATCTCTATCCACCGGAGTTTTCAATATCAAGTGATGCCACTTAATATATTATGGGGTATTAATCTCCCTTTCGAAAGGCTATCCCCCAGATAAAGGCAGGTTGCACACGTGTTNCGCACCCGTACGCCGCTCTCTCATTTCCGAAGAAACAATACCGCTCG
Name: Chryseobacterium
Qualitative: Unknown as this sample did not come from our petri dishes.
Quantitative: This species is gram negative and rod shaped.
Discussion:
Both of these samples are consistent with the data collected from the earlier labs. The white-Opaque Colony, 10^(-7) Tetracycline Free Plate bacteria was rod shaped and pink after gram staining, making it a gram negative species and consistent with both DNA fragments run.
MRS
2/26/14
Objective:
The objective of this lab was to understand invertebrates and how they differ. Also, invertebrates that were collected from the transect were analyzed to see the diversity of invertebrates in the transect. It was hypothesized that at least two different kinds of invertebrates will be observed.
Steps:
Procedure 1: Observing Acoemolates, Pseduocoelomates, and Coelomates
1)Observed the live Acoemolate Planaria and it's cross section
2)Observed the live Pseudocoelomates nematoeds and it's cross section
3)Observed the live coelomate, Annelida and it's cross section
Procedure II: Analyzing the Invertebrates Collected with the Berlese Funnel
1)Observed sample arthropods and identified them
2)Obtained prepared West Virginian samples of invertebrates, there were no invertebrates found in the Berlese Funnel
3)Compared and identified invertebrates
Procedure III: Vertebrates and Niches
1)Looked up 5 different groups of vertebrates that might inhabit the transect
2)Analyzed biotic and abiotic factors of the transect that would benefit each species
3)Construct a food web based on organisms
Data:
Procedure 1: Observing Acoemolates, Pseduocoelomates, and Coelomates
Pseduocoelomates, Nematode: The nematodes did not move much under the microscope. A few gave a twitch or two but besides this they were still.
Figure 1: Nematode Cross-section 
Coelomates, flatwords: The flatworms moved in a wave like pattern where the body would move up and down much like a sin wave in mathematics. The used this wave motion to propel themselves into the egg yolk particles in order to eat them.
Figure 2: Flatworm Cross-section 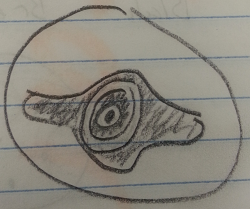
Coelomates, Earthworm: The flat worms moved in a sliding fashion. They would push the front half of their body and then slide the back half forward. The earthworms were able to move their heads in a fashion that was independent of the rest of their body.
Figure 3: Earthworm Corss-section 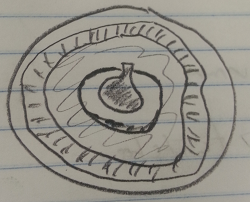
Procedure II: Analyzing the Invertebrates Collected with the Berlese Funnel
Sample Unkowns:
Station 1- Insects (some kind of bee)
Station 2- Centipede
Station 3- Crustaceans
Station 4- Arachnids
Station 5- Millipedes
Table 1: Invertebrates from WV Transect
Organism From WV Transect
Organism From WV Transect Organism Length (from internet) Brief Description of organism Flea 1.5 mm–3.3 mm 2 antenna, six legs and appears to have 2 body segments House Fly 5-7 mm 2 wings, 2 body segments Fruit Fly 3-4 mm 2 wings, one body segment Biting Lice 1-4 mm 4 legs, no antenna, 2 body segments, no wings Termite 3.5-5 mm Soft bodies, white, no wings
Procedure III: Vertebrates and Niches
N/A
Results:
Procedure I: Observing Acoemolates, Pseduocoelomates, and Coelomates
From the cross sections it can be determined that the more complex the body of the organism the more complex the movement. The flatworms had the most simple body as they did not have a coelom. They also had the simplest movement as they were only able to make wave motions using their entire body. The earthworms by contrast had an advanced coelom and the most advanced movement as they were able to move different parts of their body in different directions. It was difficult to tell about the nematodes as they did not move besides the occasional twitch when they were observed.
Procedure II: Analyzing the Invertebrates Collected with the Berlese Funnel
It should be pointed out that the Berlese Funnel from the transect did not deposit any invertebrates. It is assumed that since the leaf liter was collected on a day of heavy rain that the rain washed away all the organisms. To compensate for this samples from West Virginia were used instead.
The sizes of the samples ranged from roughly 1 mm to roughly 7 mm. The largest orgnaims observed was the house fly at around 7 mm while the smallest organism was the lice at about 1 mm. All of these organisms are arthropods. While other groups of organisms may exist in Berlese funnel leaf litter these were the only organisms sampled.
While there was some diversity among the invertebrates collected, more was expected. All the organisms observed were arthropods, no spiders or mites were observed. This may be due to the small sample size as only 5 organisms were observed. To get a better estimates of the diversity more invertebrests should be analyzed.
Procedure III:
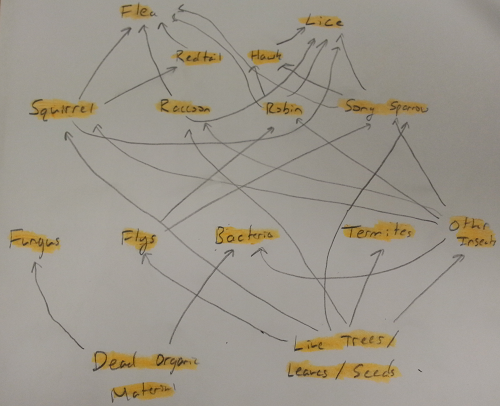
Five invertebrates who might inhabit the transect include the song sparrow, robins, raccoon, eastern gray squirrels, and red tail hawks. While none of these animals were directly observed at the transect they have been seen around campus or are known to live within Washington DC.
The song sparrows come from the Chordata phylum, Aves class, Passeriformes order, Emberizidea family, Melospiza genus, and melodia species. The songs sparrows are a a very common sparrow in the US who prefer low, dense vegetation and open ground for feeding. They are carnivores, eating small insects. Song sparrows are common in cities such as DC. While they were not seen directly it is assumed that song sparrows would like the marshy cattail area of the transect as there is both food in the from of insects and hiding spaces from predators among the reeds. (http://www.allaboutbirds.org/guide/song_sparrow/id)
The robin comes from the Chordata phylum, Aves Class, Passeriformes Order, Turidae Family, Turdus Genus, and migratorius species. The robin are a very common trush species, found throughout North America. The robin is a very versatile animal, able to live forests, tundra, plains, yards, and fields to name a few. While not seen in the transect robins have been observed on campus. In the transect the robin would most likely use it to feed among the grassy area or the marshes to eat insects. If the plants grow any berries, while not observed in the transect, the robin could eat them too. The robin may even be able to build a nest among the redtwig dogwoods but this is unlikely. Robins are not bothered by human activity so the large amount of foot traffic near the transect could deter other species, giving robins an advantage. (http://www.allaboutbirds.org/guide/american_robin/id)
Red tail hawks come from chordata phylum, Aves Class, Falconiformes Order, Accipitridea family, Buteo Genus, and jamaicensis species. The red tailed hawk is a common bird of prey found throughout all of North America and as such can adapt to any area, even urban areas like DC. While not seen at the transect a bird of prey resembling the red tail hawk was seen by the author on campus They feed on many different kinds of creatures, such as mice, voles, amphibians, squirrels, and small birds. The area around the transect make ideal hunting ground for red tail hawks as they can hunt by sitting upon a high location, such as Kogod or Katzen, and then swoop down onto their prey in the grassy area of the transect. (http://www.allaboutbirds.org/guide/red-tailed_hawk/id)
Raccoons come from the Chordata phylum, Mammalia Class, Carnivora Order, Procyonidae Family, Procyon Genus, and lotor species. The raccoon is found throughout North America and has been introduced into parts of Europe and Asia. While not seen in the transect raccoons have been spotted on campus by the author. Raccoons are able to live in a wide variety of habitats, including urban areas. They are not very picky eaters and will dine on small animals, plants, or even human trash. The area of the transect would make an ideal hunting ground for the raccoon. Small insects or other invertebrates can be located in the marshy areas and birds may nest among the redtwig doogwoods allowing the raccoon to eat eggs. Additionally, as the transect is on a college campus there is bound to be some human food waste that the raccoon could feast upon. (http://animals.nationalgeographic.com/animals/mammals/raccoon/)
The Eastern Gray squirrel comes from the Chordata phylum, Mammalia Class, Rodentia Order, Sciuridea Family, Sciurus Genus, and carolinensis species. The gray squirrel is found all along the east coast of North America. The gray squirrel was not seen at the transect but they are very common on campus. While the squirrel prefers forests and trees they can live in urban areas due to the trees humans plant. The gray squirrel mainly lives in tall deciduous trees such as oak or hickory. Gray squirrels mainly feed upon nuts, seeds, some plants, and small insects. The gray squirrel would not make the transect home as it lacks large trees for them to live in but it might come from time to time to feed upon the seeds, nuts, and insects growing in the marshy part of the transect. (http://www.fcps.edu/islandcreekes/ecology/eastern_gray_squirrel.htm)
From this information a crude food web was created using organisms from the transect.
MRS
2/26/14
Lab 4: Plantae and Fungi
Objective:
The objective of this lab was to understand the characteristics and diversity of plants and fungi. It is predicted that the transect will provide a variety of different vascularzations and cotyldeons.
Steps:
Procedure 1: Collecting five plant samples form the transect
1)Went out to transect
2)Collected a leaf litter sample of about 500g
3)Collected five separate plants samples including but not limited to leaves, seeds, branches, flowers, etc.
Procedure II: Plant Vascularzation
1)Observed the vascularzation of sample plants to get an understanding of plant vascularzation
2)Observed the vascularzatin of the plants from the transect
Procedure III: Plant Specialization
1)Observed the leaves from the sample plants to get an understanding of leaf specialization
2)Observed the leaves from the plants from the transect
Procedure IV: Plant Reproduction
1)Observed the reproductive traits of sample plants to get an understanding of plant reproduction
2)Observed the reproductive traits of the plants from the transect
Procedure V: Observing Fungi
1)Observed fungi samples under dissecting microscope
Procedure VI: Setting up the Berlese Funnel to Collect Invertebrates
1)Poured about 25 ml of 50:50 ethanol/water solution into a large centrifuge tube
2)Taped a piece of screening to the bottom of a funnel
3)Positioned funnel over tube and put leaf litter from transect into funnel
4)Covered funnel with foil and put a lamp light right over the funnel
5)Left the setup alone for a week
Data:
Procedures I-IV
Plant 1:
This plant is a tall reed of some sort with long, thin blade like leaves (figure 1). The plant has a long center stalk that was about should height from which everything grows off of. The stalk and leaves are a dull brown, most likely because the plant is dead due to it being winter. The leaves were only about as wide as a pencil but were almost as tall as the center stalk. There is a mud brown fuzzy cylinder near the top of the stalk. While the roots/stem of the this plant were not analyzed based on it's height it is assumed that this plant has vascular tissue as water and nutrients would need to be transported from the top to the bottom of the plant and vice versa. As seen in Figure 1 the leaves of this plant are long and thin and grown from bottom to middle of the stalk. The leaves do not appear very numerous, only a few grow from the stalk. Upon further investigation of the fuzzy cylinder (figure 1 and 2) it was discovered that this was a tube of very fine seeds. It appears that these seeds would be dispersed by blowing in the wind. From the plant leaves it was determined that the plant was monocot as the leaves had parallel veins. These plants are located in the wet and marshy area of center of the transect.
Plant 2:
This plant was tall, overshadowing plant 1. The plant had a bare stalk but a bushy head full of either leaves, buds, or small flowers, all of them brown and withered. While the roots of the plant were not analyzed it is assumed that this plant has vascular tissues as water and nutrients would need to be transported up and down the plant. The brown leaves on this plant are incredibly small, they are about the length of a finer nail and about as wide as two toothpicks (Figure 4 and 5). The leaves are interspersed among the small buds all along the branches of the plant (figures 4 and 5). Upon further investigation of the leaves it was determined that the small buds were little flowers indicating that the plant is an angiosperm (figure 5). It was determined from the leaves with the parallel veins and not the flowers that the plant was a monocot as the flower were too small and shriveled to observe the petal pattern. No seeds were observed. This plant was located in the same marshy area as plant 1.
Plant 3:
This plant was a shrub or small tree with red bark that grew to about chest height(figure 6). While the roots of the plant were not analyzed it is assumed that this plant has vascular tissues as water and nutrients would need to be transported up and down the plant. A few withered leaves appeared on the branches(figure 6 and 8). They were a dark green, oval shaped, and about the size of two thumbs together. A central vein went down the leaves,with a network of veins leading off indicating that this plant is dicot (figure 7) . While only a few leaves remained near the tips of the branches, markings along the bark indicated locations where other leaves could have once been all along the branches (figure 8). A few small brown buds were observed as the only reproductive parts of the plant, figure 6. This plant sample was located on the left hand side of the transect near the edge of the grass/rock line.
Plant 4:
This plant was a very low lying plant that grew among the rocks (figure 9). The roots of this plant were observed and it is thought that this plant has vascular tissue. The leaves of this plant are bright green and are about half a finger long (figure 10). The plant is a dicot as there is a central vein on the leaf that other veins branch off of (figure 9). It was hard to determine if there was a stem on this plant, the leaves grew so thick and close together that it was like having a ball of leaves (figure 10). No flowers, buds, or spores were observed on this plant. This plant was located among the rocks to the right on the marshy area near the border between the grass and the rocks.
Plant 5:
This plant is a grass, the normal kind observed on the ground, growing only about two inches high(figure 12). The roots of this plant were observed and it is thought that this plant has vascular tissue. The leaves of this plant are green and very narrow, about half a pencil width. The leaves/blades of grass are not that close to each other, it is almost like each blade is its own plant. The plant is a monocot as the veins in the leaf run in a parallel direction. No flowers, buds, or spores were observed on this plant. The plant was located in the grassy area at the top of the transect.
Figure 12: Plant 5 at Transect 
Procedure V: Observing Fungi
This sample had white fibers and small black dots on the fibers.
Procedure VI:
No Data was collected
Results
Procedure I-IV
Plant 1 was determined to be a cattail. Cattails do have flowers so they are a monocot angiosperms. The scientific name for cattails is T. angustifolia (http://www.cattails.info/). Plant 2 was determined to be a monocot angiosperm. The exact genera for this species was unable to be determined from the information provided. Plant 3 was determined to be a Redtwig dogwood or Cornus alba (http://www.finegardening.com/plantguide/cornus-alba-elegantissima-red-twig-dogwood.aspx). Redtwig dogwoods are dicot angiosperms. Plant 4 was unable to be identified but it is assumed to be a dicot angiosperm. This is because based on the plant and it's structure it seems highly unlikely to be a gymnosperm and the leaf structure did not resemble those of ferns. Plant 5 is known to be a type of grass as it was torn out of the lawn. Therefore, plant 5 is a monocot angiosperm (http://en.wikipedia.org/wiki/Poaceae).
The prediction was validated in that we did find a mixture of monocot and dicot plants. However, it was also discredited because all the plants sampled were vascular plants. This may indicate that the transect is very complex and that organisms need a high level of order and specialization in order to survive.
Procedure V: Observing Fungi
Fungi sporangia are small, black balls formed from the white hyphae filaments. They contain spores which are released once the sporangia open up. Based upon figure 14 it is theorized that the fungus in question is black bread mold, a kind of zygomycota. It is believed to be a fungus as it has wispy white filaments and small black dots. These are the hyphae and sorangia, structures only found in fungi.
MRS
2/16/14
Lab 3: Microbiology and Identifying Bacteria with DNA
Objective:
The objective of this lab was to learn the differences between bacteria based on physical shape and the presence, or lack of, an outer peptidoglycan layer. Also, this lab explores antibiotics and how they could affect bacterial growth in an agar plate. It is predicted that there will be a diverse group of bacteria, as in more than 2 shapes and a mixture of gram negative or positive colonies, and that there will be less colonies on the agar plates with tetracycline.
Steps:
Procedure I: Quantifying and Observing Bacteria
1)Observed hay infusion for signs of change in smell or appearance
2)Counted number of colonies formed on agar plates. For small number of colonies they were outright counted, However, for plates with large number of colonies the plate was divided up into sections and the number of colonies found in a section multiplied by the number of sections to find the total number of colonies on a plate.
Procedure II: Antibiotic Resistance
1)Compared differences between plates with the same concentrations but with or without tetracycline present.
2)Found information about tetracycline and the types of bacteria sensitive to this antibiotic
Procedure III: Bacteria Cell Morphology Observations
1)Observed prepared slides of bacillus, coccus, and spirillum shaped bacteria
2)Sterilized 3 loops
3)Collected a pink colony from the 10^(-3) tetracycline plate, an white-opaque colony from the 10^(-7) tetracycline free plate, and an orange colony from the 10^(-5) tetracycline free plate. Made a wet mount of each colony sample
4)Observed bacteria colonies under the microscope. This was done to determine the size and shape of the bacteria.
5)Made gram stain for the bacteria by first drying the slide under a flame for several seconds. Then, the bacteria was stained with crystal violet. The crystal violet was washed off with water and then covered with Gram's iodine mordant before being washed with 95% alcohol. Finally, the bacteria was covered with safranin strips and then washed.
6)The bacteria samples post gram stain treatment were observed under the microscope to determine size and shape.
Procedure IV: PCR Preparation
1)Selected the black bacteria colony from 10^(-5) tetracycline free plate and the green colony from 10^(-3)tetracycline plate for PCR sequencing.
2)Selected colonies were transferred into 100 microliters of water in a sterile tube and incubated for 10 minutes in 100 degree Celsius water bath.
3)Tubes were centrifuged and 5 microliters of the supernatant removed for the PCR reaction
Data:
Procedure I: Quantifying and Observing Bacteria
Hay infusion: The hay infusion's water was much darker than before and some appeared to have evaporated as there was residue on the wall of the container. The water also appeared to be more homogeneous, there was only a slight discoloration of the water near the bottom among the organic debris. There still appeared to be algae like growth on the top of the surface. The container smelled less than it did last time.
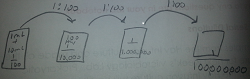
Table 1: 100-Fold Serial Dilution Results
Dilution (plate label) Agar Colonies Counted Conversion Factor Colonies/ml 10^-3 Nutrient 1940 x 10^3 1940 x 10^3 10^-5 Nutrient 150 x 10^5 150 x 10^5 10^-7 Nutrient 6 x 10^7 6 x 10^7 10^-9 Nutrient 0 x 10^9 0 10^-3 Nutrient + tet 50 x 10^3 50 x 10^3 10^-5 Nutrient + tet 3 x 10^5 3 x 10^5 10^-7 Nutrient + tet 0 x 10^7 0
It is important to note that there was fungal growth on the 10^-3 and 10^-7 tetracycline plates
Procedure II:Antibiotic Resistance
There appeared to be several differences between the normal agar plates and the ones with tetracycline. First, there were several fungus spores on the 10^-3 and 10^-7 tetracycline plates. The colonies on the tetracycline plates also appeared to be smaller and less numerous than those on the normal nutrient plates. The orange and white bacteria appeared on both kinds of plates while the clear bacteria appeared on the tetracycline free plates only. There was a solitary pink colony on the 10^-3 tetracycline plate.
Procedure III: Bacteria Cell Morphology Observations
Pink Colony, 10^(-3) Tetracycline Plate: This pink colony appeared tear shaped, convex, and very smooth. On the first microscopic observation the bacteria appeared very small, round, and squished together with no movement. The bacteria was guessed to be a cocci species. After the gram-stain test it was found that this bacteria is purple, making it gram positive, and spherical with clusters. This meant the bacteria is a definitely a Bacilli bacteria, specifically staphylococcus.
Figure 1: Pink Colony, 10^(-3) Tetracycline Plate Pre-Gram Stain 40x 
Figure 2: Pink Colony, 10^(-3) Tetracycline Plate Post-Gram Stain 40x 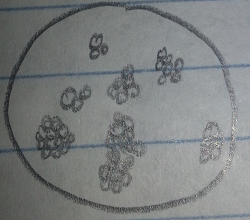
Orange Colony, 10^(-5) Tetracycline Free Plate: This orange colony was circular, convex, and smooth. On the first microscopic observation the bacteria appeared very, very small and squished together. There was no movement noticed but a few squiggle shaped objects were observed among the small bacteria. The bacteria was guessed to be two separate bacteria species, a cocci and a spirilum. After the gram-stain test it was found that the bacteria was spherical and pink, making it gram negative. The odd squiggles noticed in the pre-gram stain wash also disappeared. From the evidence gathered in the gram stain observation it was determined that the bacteria was a cocci, possibly a staphylococcus.
Figure 3: Orange Colony, 10^(-5) Tetracycline Free Plate Pre-Gram Stain 40x 
Figure 4: Orange Colony, 10^(-5) Tetracycline Free Plate Post-Gram Stain 40x 
White-Opaque Colony, 10^(-7) Tetracycline Free Plate: This white-opaque colony bacteria was very small, squished together, and had no movement. The bacteria was guessed to be a cocci species based on this evidence. After the gram-stain test it was found that the bacteria was rod shaped and pink, making it a gram negative species. This new evidence suggested that the bacteria was a bacilli, specifically diplobacilli.
Figure 5: White-Opaque Colony, 10^(-7) Tetracycline Free Plate Pre-Gram Stain 40x 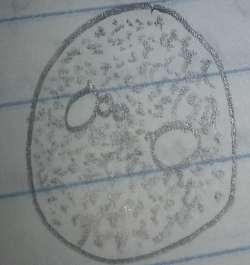
Figure 6: White-Opaque Colony, 10^(-7) Tetracycline Free Plate Post-Gram Stain 40x 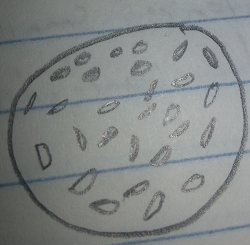
Procedure IV: PCR Preparation
No Data was recorded for this procedure
Conclusions:
Procedure I: Quantifying and Observing Bacteria
The change in smell of the hay infusion could be because the top was left open on the jar allowing for more oxygen to get in. This would allow more aerobic organisms to flourish rather than anaerobic organisms that could smell worse. Also, the availability of nutrients could have shifted as all the food originally in the water was consumed. This might allow more producers to thrive and clean up the smell and color of the water.
It is predicted that if Archea were placed on the nutrient agar plates they would not grow. Archaea grow in extreme conditions and would not flourish under such mild conditions. They would either need temperatures too high, temperatures too low, salt content too high, or any other kind of specific range that this lab cannot provide. Also, competition from normal bacteria would prevent the Archaea from gathering the nutrients required for growth and lead to the death of all Archaea as the normal bacteria are much better suited for this environment.
Procedure II:Antibiotic Resistance
The tetracycline definitely had a negative influence on the size difference of the bacterial colonies and their frequency, backing up our original prediction. The tetracycline free plates were much more populated indicating that some of the bacteria was killed off by the tetracycline. As only the 10^-3 and 10^-5 plates grew bacteria we can only compare their data with the plates of equivalent dilution. The 10^-3 tetracycline free plate grew about 40 times as much bacteria as compared to the plate with the antibiotic and 10^-5 plate grew about 50 times as much bacteria compared to its antibiotic free plate. Thus, bacteria are able to live on tetracycline plates, just at a reduced rate of growth.
Based on the colors it appears that the clear colonies will only grow on tetracycline free plate while the pink colonywill only grow in the presence of tetracycline, specifically the 10^-3 plate. The orange and white-opaque bacteria appeared on both plates.
As for fungi, they only appeared on the tetracycline plates. This is not surprising as antibiotics do not affect fungus as they only kill bacteria. Since there were fewer bacteria present than in a normal plate the fungus had an advantage and were able to flourish. On the normal plates the bacteria had the advantage and were able to kick out the fungi.
The antibiotic tetracycline works by inhibiting the synthesis of proteins. It does this by binding to the small 30s ribosomal subunit found in bacteria. When tetracycline binds to the ribosome it prevents the synthesis of proteins by blocking tRNA from coming in and binding to the ribosome. Thus, while tetracycline does not outright kill the bacteria it prevents it from producing proteins which will cause the cell to die when vital proteins do not get synthesized. Tetracycline is able to affect both gram negative and gram positive bacteria as both outer layers allow the tetracycline to enter, either though porins or diffusion.
Procedure III: Bacteria Cell Morphology Observations
The bacteria were very difficult to observe without the gram staining. The bacteria were so squished together it was hard to tell if it was part of the actual arrangement of the bacteria species or from just the density of the bacteria. With the washing however enough bacteria was cleared away to allow better observation of the bacteria. This even changed to shape seen as with the white-opaque bacteria they were originally thought to be spherical when in fact there were rod shaped. Also, the gram staining colored all the cells making them easier to see than without any kind of dye.
Our original hypothesis was also proven as we saw both gram positive and gram negative bacterial strains along with two kinds of bacteria species. The two that were the same, cocci staphylococcus, even differed on how they were gram negative or positive indicating that even among bacterial species there is great diversity.
Procedure IV: PCR Preparation
It is assumed that there will be some difference in the 16s RNA gene between the two bacteria species. Many bacteria groups have different sequences for this gene so it is predicted that two genes of different length will be discovered.
MRS
2/9/14
Lab 2: Identifying Algae and Protists
Objective:
Learn how to identify algae and protists through physical characteristics. After the acquisition of necessary identification skills, two different samples were taken from the hay infusion prepared in the previous lab and analyzed for protists and algae. It was hypothesized that there would be more aerobic producers near the sample from the top of the jar as the organisms would have access to sunlight and oxygen in addition to several consumers that would eat the producers. Anaerobic consumers would be found more in the sample from the bottom of the jar where there was lots of dead organic material to eat, less oxygen, and a little sunlight. With so much food on the bottom and little sunlight/oxygen there would be no use for there to be many producers. Once the protists and algae were discovered they were then identified using a dichotomous key.
Steps:
Procedure I: How to Use a Dichotomous Key
1)Made a wet mount of known organisms and observed them at various magnifications under the microscope (4x, 10x, and 40x)
2)Measured organism
3)Identified organism using Dichotomous Key
4) Repeated with second organism
Procedure II: Hay Infusion Culture Observations
1)Obtained hay infusion created in last lab
2)Observed hay infusion for appearance, smell, and other physical characteristics
3)Obtained samples hay infusion water from area close to the surface of the water and from the bottom in order to make wet mounts
4)Observed, measured, and identified three separate organism from each location (top or bottom of the jar)
Procedure III: Preparing and Plating Serial Dilutions
1)Obtained and labeled four normal agar plates 10^(-3), 10^(-5), 10^(-7), and 10^(-9). Also obtained and labeled four tetracycline coated agar plates Tet 10^(-3), 10^(-5), 10^(-7), 10^(-9).
2)Obtained four tubes of 10mls sterile broth and labeled 2,4,6,8.
3)Swirled Hay Infusion
4)Obtained 100 microliters of infusion fluid and added to test tube 2
5)Made serial dilutions with tubes 2,4,6, and 8 using 100 micoliter increments of liquid
6)After dilution, took 100 microliters from each test tube and added it to the 10^(test tube number + 1) plate for both tet and non tet
Data:
Procedure I: How to Use a Dichotomous Key
Example Organism 1: The first organism was about 35-30 micometers long. It was determined to be a Peranema after consulting the Dichotomous Key.
Example Organism 2: The first organism was bout 5 micometers long. It was determined to be a Euglena after consulting the Dichotomous Key.
Procedure II: Hay Infusion Culture Observations
Observations of Hay Infusion- The infusion smelled like mold or old seafood, an overall unpleasant odor. There was some algae or other plant growth on the surface of the water and under it creating a whitish film. Overall the water near the top was very cloudy. There were two small sprouts growing out of seeds that were on twigs above the water surface. The middle layer of water was transparent, but colored like weak tea. The bottom of the jar contained lots of sediment from the fallen leaves and other organic debris.
Samples- Two samples were taken from the hay infusion, one near the top of the water line right under the white film and the other at the bottom among the sediment.
Sample From the Top of the Jar
Top Organism 1: This organisms was motile and was bout 5-6 micrometers long. From its features it was determined to be a Euplotes, a nonphotosynthesizing protozoa.
Figure 3: Top Organism 1, Euplotes 
Top Organism 2: This organism was motile and only about 2 micrometers long. From its features it was determined to be a Colpidium, a nonphotosynthesizing protozoa.
Figure 4: Top Organism 2, Colpidum 
Top Organism 3: This organism was motile and about 7 micrometers long. From its features it was determined to be a Colpidium, a nonphotosynthesizing protozoa.
Figure 5: Top Organism 3, Colpidum 
Sample From the Bottom of the Jar
Bottom Organism 1: This organisms was gray, motile, and about 2.5micrometers long. From its features it was determined to be a Colpidium, a nonphotosynthesizing protozoa.
Figure 6: Bottom Organism 1, Colpidium 
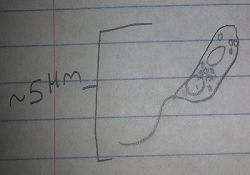
Bottom Organism 2: This organism was green, motile, and about 1.5 micometers long. From its features it was determined to be a Chlamydomonas, a photosynthesizing green algae.
Figure 7: Bottom Organism 2, Chlamydomonas 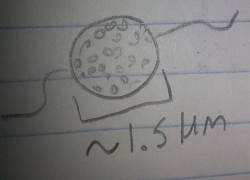
Bottom Organism 3: This organism was gray, motile, and about 3.25 micometers long. From its features it was determined to be an Ameoba, a nonphotosythesizing protozoa.
Figure 8: Bottom Organism 3, Ameoba 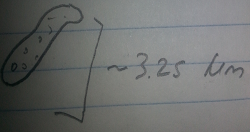
Procedure III: Preparing and Plating Serial Dilutions
Figure 9: Serial Dilution
Conclusion:
The original hypothesis was wrong, the only producer observed in the sample came from the bottom of the jar, not the top. This may be because the white film/cloudiness of the water near the top was due to macroscopic organisms, not microscopic ones like the chlamydomans. Then, the consumers, such as the colpidium, would eat the producers. On the bottom however, there may not actually be as much nutrition or competition for producers and as such the chlamydomans can capture a better niche. Thus, in the long term there may be selective pressures for more producers once the available nutrition for consumers runs low.
If the hay infusion was allowed to continue on for longer I would expect to see more producers and less consumers. This is because the consumers will eat all the available nutrients from the infusion and more producers would be needed to provide food. I would also expect larger and more complex organisms to grow. This is because as the hay infusion continues the organisms that are will suited to the environment will grow larger and start to dominate over the general organisms. Thus, the organisms in the niches will become more complex and suited for that role than the earlier generalists.
While no plant matter was removed for inspection, it is hypothesized that more consumers would be found near them. This is because the plants take in all the nutrients other producers, such as chlamydomonas, would use. This leaves only the consumers to eat the plants and its byproducts by the plants.
From the Freedman text organisms must 1) acquire and use energy 2)are made up of cells 3)process information 4)are capable of replication 5)are a product of evolution. The second Colpidium, Top Organism 3, follows all these rules. The organism moved around using its cilia, a process that requires the use of energy proving point 1. It also appeared to be made of a single cell, fulfilling number 2 and 3. To be made from a cell the Colpidium must be made of DNA meaning it has a hereditary path for information, aka DNA. While no replication was observed, multiple Colpidium of the same species were located meaning that the organism must replicate in some manner, number 4. The final point is hard to prove after just several minutes of observation, but the fact that another Colpidium species was observed gives evidence to evolution. The two species differed in both size and shape, with the second one having a cleft on one side of its body. If both species came from one common ancestor they must have diverged somewhere along the way.
While the results from the serial dilution will not be known until the next lab, it is assumed that less bacteria will grow on the plates with tetracycline as tetracycline is an antibiotic and will kill most bacteria. Also, less bacteria will grow on the plates with lesser concentrations as there are less different bacteria to form colonies with.
MRS
2/6/14, lab 1 notes
Great characterization of your transect! Make sure you include pics of lab 1 and lab 2 by Sunday. Also, start working on building a map of your transect to detail your land and where your samples are taken from. We will talk about this more Wednesday.
AP
1/30/14
Lab 1: Biological Life at AU
Objective: Observe a transect of the AU campus with regards to its biotic, abiotic, and other general features. By looking at a 20 ft by ft square of land here at AU we can observe how organisms and the environment interact within a specific section of land.
Steps:
1)Walked to assigned plot of land
2)Observed and recorded the biotic, abiotic, and other general features of the 20 by 20 foot plot of land
3)Took a top soil and ground vegetation sample for a hay infusion culture
4)Made a hay infusion culture by mixing 10 to 12 grams of the soil sample and placing it in a plastic jar with 500 mls of deerpark water. Then, 0.1 gm dried milk was added and gently mixed for about 10 seconds. The jar was capped and set aside to culture.
Data:
The plot observed was located next to Kogod alongside the footpath in the grass/vegetation area along Massachusetts Avenue.
Abiotic components: About half the plot was in shade at that time of the day, about 1:15pm. The soil near the cattail was very wet and damp, the area appears to be a runoff area as there is a drain located right outside the limits of the plot. Several different types of rocks appeared in the area. When facing Massachusetts Avenue there was a patch of very small, only a few inches across, rocks that were surrounding two larger boulders on the lower left hand side of the plot. The area closer to Kogod will be the lower side, the side closer to Massachusetts Avenue the upper side. To the right of this small stone patch was another stone patch, this time with rocks about the size of two or three fists. Three large, flat boulders appeared in this area. There is lots of exposed soil near the stone patches.
Biotic Components: There is a triangle of grass in the upper left side of the plot that takes up about ¼ of the plot. There is very little besides grass in this area in terms of life. On the very left border of the plot and in the middle of the plot on the grass/dirt border there are two red barked shrubs that did not have any leaves. On the middle to the right hand side of the plot in the stone/dirt area there is a small thicket of cattails. Among the cattails there are a few small plants that may be weeds. To the right of the cattails are a several very small plants that hug the ground. On several of the rocks either a moss or algae is growing, likely due to the large amounts of water the area gets. Lots of organic matter, such as dropped leaves, sticks, or other dead plant material, litter the ground in the stone area and among the cattails.
Figure 1: Picture of Transect
Conclusion:
In short, most of the plot is either grass or a field of stones of various sizes. Two red shrubs and a thicket of cattails dominate the biological scene. The area appears to get lots of water due to the slope of the surrounding land leading runoff into the plot. Other studies will be performed using the hay culture created in this lab to observe the microorganisms located in the plot. Also, the plot will be revisited later to see if there are any changes to it.
MRS
11/22/14
Entered text successfully on this date
MRS