User:Lindenb/Notebook/UMR915/20110511
From OpenWetWare

Integragen
analyse SNP_het (het. composite)
#remove rs
#only keep the 'SNP_het'
#remove the low qualities
#remove SNP_het*
#only the non-synonymous or stop
#remove DNA & prot sequences
#add chrom_position flag
#sort
gunzip -c AllChrom.exome.snp.pool.new.annotation.gz |\
awk -F ' ' '{if(substr($4,1,2)!="rs") print;}' |\
awk -F ' ' '{if(index($19,"douteux")==0) print;}' |\
awk -F ' ' '{if(index($19,"_het")!=0) print;}' |\
awk -F ' ' '{if(index($26,"nonsense")!=0 || index($26,"missense")!=0) print;}' |\
cut -d ' ' -f 1-27 |\
awk -F ' ' '{printf("%s_%s\t%s\n",$2,$1,$0);}' |\
sort -t ' ' -k1,1 > _jeter1.txt
#get all distinct chrom_pos in file
cut -d ' ' -f 1 _jeter1.txt | sort -t ' ' -k1,1 | uniq > _jeter2.txt
#extract wild exome
#keep chrom,position
#add chrom_position flag
#sort
gunzip -c AllChrom.exome.snp.u2437.new.annotation.gz |\
cut -d ' ' -f 1,2 |\
awk -F ' ' '{printf("%s_%s\n",$2,$1);}' |\
sort -t ' ' -k 1,1 | uniq > _jeter3.txt
#get [m] chrom_pos not in [+] chrom_pos set
comm -2 -3 _jeter2.txt _jeter3.txt > _jeter4.txt
#join uniq [m] chrom_pos & mutated data
#remove chrom_pos
#order by gene
join -t ' ' --check-order -1 1 -2 1 _jeter1.txt _jeter4.txt|\
cut -d ' ' -f 2- |\
sort -t ' ' -k 20 > _jeter5.txt
#join to self using key= "gene name"
#only keep if first mutation in same gene/chromosome and pos1< pos2
#keep some columns
join -t ' ' -j 20 _jeter5.txt _jeter5.txt |\
awk -F ' ' '{if($3==$29 && int($2) < int($28) ) print;}' |\
cut -d ' ' -f 1,2,3,20,26,28,46,52 > _jeter6.txt
#extract gene names
cut -d ' ' -f 1 _jeter6.txt | sort | uniq
rm _jeter[12345].txt
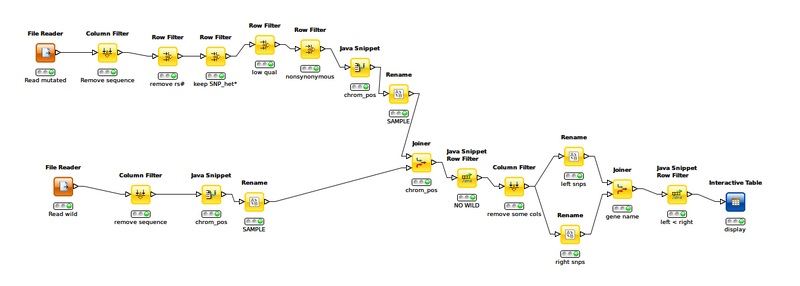
Knime equivalent
Slideshow
<html>
<a href="http://www.slideshare.net/lindenb/analyzing-exome-data-with-knime" title="Analyzing Exome Data with KNIME">Analyzing Exome Data with KNIME</a> <iframe src="http://www.slideshare.net/slideshow/embed_code/7923093" width="425" height="355" frameborder="0" marginwidth="0" marginheight="0" scrolling="no"></iframe>
View more <a href="http://www.slideshare.net/">presentations</a> from <a href="http://www.slideshare.net/lindenb">Pierre Lindenbaum</a>
</html>
SNP_diff
#remove rs
#in gene
#remove the low qualities
#keep SNP_diff
#only the non-synonymous or stop
#remove DNA & prot sequences
#order by GENE
gunzip -c AllChrom.exome.snp.pool.new.annotation.gz |\
awk -F ' ' '{if(substr($4,1,2)!="rs") print;}' |\
awk -F ' ' '{if($20!="") print;}' |\
awk -F ' ' '{if(index($19,"douteux")==0) print;}' |\
awk -F ' ' '{if(index($19,"_diff")!=0) print;}' |\
awk -F ' ' '{if(index($26,"nonsense")!=0 || index($26,"missense")!=0) print;}' |\
cut -d ' ' -f 1-27 |\
sort -t ' ' -k20,20 > _jeter1.txt
#extract wild exome
#remove rs
#remove SNP_diff
#in gene
#order by gene
gunzip -c AllChrom.exome.snp.u2437.new.annotation.gz |\
awk -F ' ' '{if(substr($4,1,2)!="rs") print;}' |\
awk -F ' ' '{if(index($19,"douteux")==0) print;}' |\
awk -F ' ' '{if(index($19,"_diff")==0) print;}' |\
awk -F ' ' '{if($20!="") print;}' |\
cut -d ' ' -f 1-27 |\
sort -t ' ' -k20,20 > _jeter3.txt
#join wild & mutated data by gene
#check wild sample has no mutation in the pair of mutated snps
#remove wild data
join -t ' ' -1 20 -2 20 _jeter1.txt _jeter3.txt |\
awk -F ' ' '{if($3==$29 && int($2) == int($28) ) print;}' |\
cut -d ' ' -f 1 |\
sort | uniq
rm _jeter*.txt