Objective
The purpose of this experiment is to use PCR to mutate GFP so that a cysteine residue can replace the aspartic acid residue directly after the enterokinase cleavage site on the vector.
Procedure
- Two complimentary primers (listed below) containing the desired mutation were synthesized and purified.
- A sample reaction was prepared as follows:
- 5 μl of 10× reaction buffer)
- 1.25 μl of dsDNA template
- 1 μl of the forward primer
- 1 μl of the reverse primer
- 1 μl of dNTP mix
- double-distilled water (ddH2O) to a final volume of 50 μl
- At the end just before temperature cycling, 1 μl of PfuTurbo DNA polymerase and 25μl of wax were added to the sample.
- Each reaction was placed underwent specific cycling parameters, which were as follows:
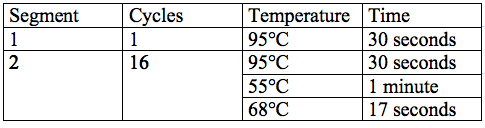
- The reaction was also cycled for 10 minutes at 72°C and for 24 hours at 4°C.
Results
- A suitable primer was researched for this experiment and determined to be:
Forward Primer:
GAT AAG GAT GAC GAT AAG TGT CGA TGG GGA TCC GAA TTC GCC
Reverse Primer:
GGC GAA TTC GGA TCC CCA TCG ACA CTT ATC GTC ATC CTT ATC
- This primer was 39 base pairs, had a GC content of 51%, and had a salt adjusted melting temperature of 78.8°C.
- The actual primer used was:
Forward Primer:
5'- GAC GAT GAC GAT AAG GAT CGA TGG GGA TCC GAA -3'
Reverse Primer:
5'- TTC GGA TCC CCA TCG ATC CTT ATC GTC ATC GTC- 3'
- This primer was 33 base pairs.
Notes
| 
