User:Fermenter User/Notebook/Pichia Workshop 20100809
|
WORK A1 Teaching:
Day -2 (Aug 9 Mon): <Dennis> 8/9/2010 (during workshop)
Thaw glycerol stock and grow 5-ul in fresh 5-ml MD media
Incubator in Fermentation Suite used
<Dennis> 8/9/2010 (before go home)
Give BMXY (yeast extract + peptone for 3.5-L x2) to Sung-Hye
Prepare:
YNB, Phosphate buffer, Glycerol, Biotin, Acid (5M Acetic Acid)
Day -1 (Aug 10 Tue): MEDIA PREPARATION DAY <Workshop> 8/10/2010 (10:00) Assemble bioreactors:
1. Calibrate pH sensors
2. 3.5-L of BMXY (Fermenter #1,#2)
5.0-L of Basal Salts (Fermenter #3):
3. 1-ml Antifoam
<Raymond> 8/10/2010 (11:50 - 12:00)
Spind down cells and resuspend in fresh 150-ml BMGY media
Incubator in Fermentation Suite used
Day 0 (Aug 11 Wed): INOCULATION DAY <Workshop> 8/11/2010 (10:18) Start Fermenter #1 software
8/11/2010 (10:20) Start Fermenter #2 software
8/11/2010 (10:22) Start Fermenter #3 software
8/11/2010 (10:30) Set Temperature
Purge Air 1 L/min at 600 rpm
Does anybody has picture of inoculation today? If so, feel free to upload at Inoculation-20100811.jpg.
Day 1 (Aug 12 Thu): <Sung-Hye> 8/12/2010 (10:00)
F2: water leakage from heat exchange water out. repaired
F1: Temp control was not working (cooling was not sufficient to maintain temp at 28.5°C)
Ice was used to bring down temp to SP and removed.
<Workshop> 8/12/2010 (12:00) Fermentation 24:00
F1 Acid(310 ml) Base(390 ml) OD(19.8) Contamination(N) Pressure(N) Change Filter(N)
F2 Acid(300 ml) Base(380 ml) OD(6.94) Contamination(N) Pressure(N) Change Filter(N)
F3 No Acid Base(400 ml) OD(0.6) Contamination(N) Pressure(N) Change Filter(N)
Day 2 (Aug 13 Fri): MeOH Induction Day 1 <Sung-Hye> 8/13/2010 (09:00 - 12:00)
Glycerol + PTM1 Salt Feed : 90 ml/hr, Final voluem: 180 ml
<Workshop> 8/13/2010 (11:00) Fermentation 48:00
F1 Acid(130 ml) Base(380 ml) OD(22.15)Contamination(N) Pressure(N) Change Filter(N)
F2 Acid(150 ml) Base(380 ml) OD(22.3) Contamination(N) Pressure(N) Change Filter(N)
F3 No Acid Base(300 ml) OD(38.2) Contamination(N) Pressure(N) Change Filter(N)
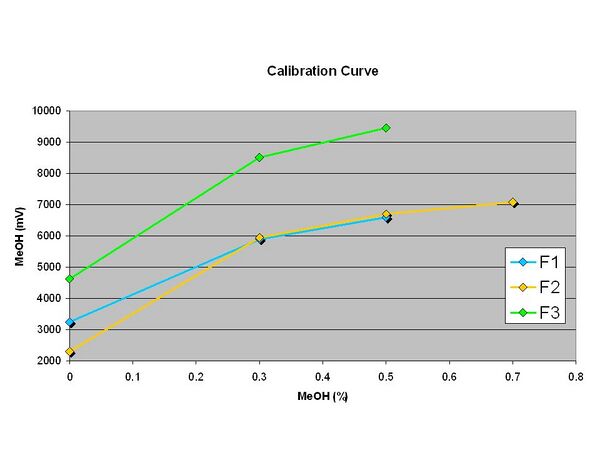
<Workshop> 8/13/2010 (12:00) MeOH sensor calibration
8/13/2010 (12:00) Fermentation 48:00 MeOH induction 00:00
Connect MeOH
7. 800 ml of MeOH: Tasfer MeOH into MeOH feeding bottles.
<Sung-Hye> 8/13/2010 (12:00)
Harvest F3 500 ml
<Sung-Hye> 8/13/2010 (15:12)
F1 Acid (110 ml) + Add (500 ml) = (610 ml)
F2 Acid (100 ml) + Add (500 ml) = (600 ml)
Day 3 (Aug 14 Sat): MeOH Induction Day 2 <Sung-Hye> 8/14/2010 (10:00)
F3: Set MeOH to 9000 mV (Max allow)
<Dennis> 8/14/2010 (12:00) Fermentation 72:00 MeOH induction 24:00
F1: Acid(590 ml) Base(390 ml) MeOH(25 ml injection) OD(22.45)
Contamination(N but I did observe one bacterium, possibly from dirty syringe?) Pressure(N) Change Filter(N)
Activity assay (4.02 units/100ul), (0.94 mg/5-L)
F2: Acid(560 ml) Base(375 ml) MeOH(790 ml) OD(20.2) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (1.03 units/100ul), (0.24 mg/5-L)
F3: NO Acid Base(275 ml) MeOH(775 ml) OD(42.25) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (6.91 units/20ul so 34.55 units/100ul), (8.08 mg/5-L) <-- WOW!
Day 4 (Aug 15 Sun): MeOH Induction Day 3 <Dennis> 8/15/2010 (12:00) Fermentation 96:00 MeOH induction 48:00
F1: Acid(560 ml) Base(390 ml) MeOH(25 ml injection) OD(23.2) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (7.20 units/100ul), (1.68 mg/5-L)
F2: Acid(540 ml) Base(375 ml) MeOH(755 ml) OD(21.3) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (1.91 units/100ul), (0.45 mg/5-L)
F3: NO Acid Base(275 ml) MeOH(755 ml) OD(44.3)
Contamination(N there were a few bacteria but not enough for me to claim contamination) Pressure(N) Change Filter(N)
Activity assay (2.132/5ul so 42.64 units/100ul), (9.98 mg/5-L)
Day 5 (Aug 16 Mon): MeOH Induction Day 4 <Workshop> 8/16/2010 (12:00) Fermentation 112:00 MeOH induction 72:00
F1: Acid(?? ml) Base(?? ml) MeOH(25 ml injection) OD(23.75) Contamination(N) Pressure(Y/N) Change Filter(Y/N)
Activity assay (13.26 units/100ul), (?? mg/5-L)
F2: Acid(525 ml) Base(380 ml) MeOH(750 ml) OD(19.5) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (3.66 units/100ul), (0.856 mg/5-L)
F3: NO Acid Base(300 ml) MeOH(750 ml) OD(41.35) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (3.74 units/5ul so 74.8 units/100ul), (17.5 mg/5-L)
(WORKSHOP ENDS HERE) Day 6 (Aug 17 Tue): MeOH Induction Day 5 <Dennis> 8/17/2010 (12:00) Fermentation 136:00 MeOH induction 96:00
F1: Acid(480 ml) Base(390 ml) MeOH(25 ml injection) OD(22.5) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (14.68 units/100ul), (3.44 mg/5-L)
F2: Acid(500 ml) Base(375 ml) MeOH(700 ml) OD(25.6) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (5.74 units/100ul), (1.34 mg/5-L)
F3: NO Acid Base(275 ml) MeOH(725 ml) OD(48.7) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (3.72 units/5ul so 74.4 units/100ul), (17.4 mg/5-L)
Day 7 (Aug 18 Wed): MeOH Induction Day 6 : HARVEST <Dennis> 8/18/2010 (12:00) Fermentation 160:00 MeOH induction 120:00
F1: Acid(460 ml) Base(390 ml) MeOH(25 ml injection) OD(25.1) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (15.91 units/100ul), (3.72 mg/5-L)
F2: Acid(500 ml) Base(375 ml) MeOH(670 ml) OD(23.85) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (4.87 units/100ul), (1.14 mg/5-L)
F3: NO Acid Base(275 ml) MeOH(705 ml) OD(46.05) Contamination(N) Pressure(N) Change Filter(N)
Activity assay (2.59 units/5ul so 51.8 units/100ul), (12.12 mg/5-L)
HARVEST
Discussion 1: ZeocinQuestion: How Zeocin works? *Sung-Hye Grieco 11:34, 11 August 2010 (EDT): Zeocin (copper chelated glycopeptide antibiotic) belongs to the bleomycin/ phleomycin family of antibiotics. It acts on mammalian, insect cells, yeast, bacteria and plants via intercalating into DNA and cleaving it, thereby causing cell death. The Sh ble gene gives resistance to Zeocin. The gene product binds to Zeocin and therefore inhibits its intercalation into the DNA.
Discussion 2: Media-relatedQuestion: Why we use MD media in the begining and then change to BMGY? *Sung-Hye Grieco 12:53, 11 August 2010 (EDT): Apparently minimal media selects for double auxotrophs ? Maybe rich media loosens the selection criteria and allows one of the picZ insertions on the diploid chromosome to be dumped ? so by selecting on MD you are pickingdouble transformants in which chromosomal integration has taken place on both genomes ? Just a thought ... (Raj)*Sung-Hye Grieco 13:54, 17 August 2010 (EDT): Discussion 3: Media-related Question: Why would Fermenter #3 (the Defined Medium) have more optimal growth than Fermenter #2
again? Fermenter #2 is predicted to perform better than #1 because it has the feedback.*Shiau-Yun Tang 20:27, 11 August 2010 (EDT):
Becasue Fermentation strategy using define medium (Fermenter #3) includs Fed-Batch with Glycerol. If Glycerol is fed in growth-limiting rate, it increases biomass and also primes methanol-utilization pathway enzymes before induction. Feed rate and feed time will depend on the desired cell density and the end of the glycerol fed batch phase, which may vary from 150 to 450 WCW/L, and should be optimized based on teh production time course. Reference: Pichia Protocols, 2nd edition, p47 *Sung-Hye Grieco 19:12, 13 August 2010 (EDT):
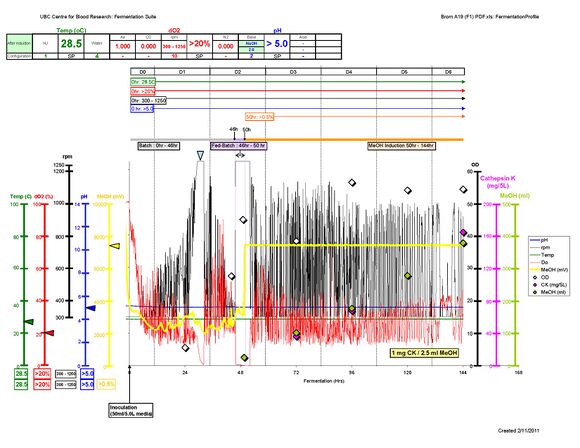
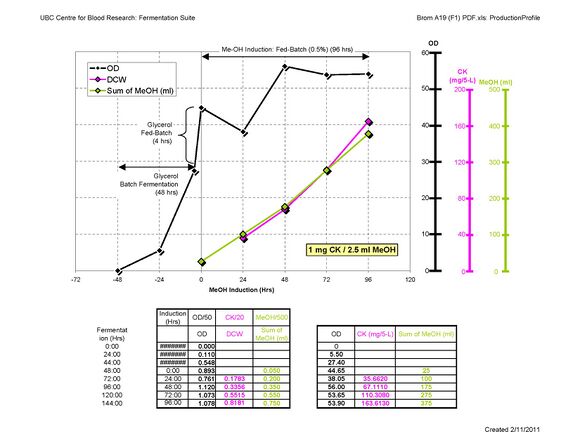
Follow up:Recently, Project BROM-A19, produced a record-breaking amount of human Cathepsin K! We procuded roughly 160 mg from 5-L Defined media. The protocol was same but with slight modification of base and it seems that timing of Fed-Batch plaied a quite important role. *Sung-Hye Grieco 17:16, 15 February 2011 (EST):
AnnouncementWorkshop Time Schedule ReferencesInvitrogen Pichia Expression Kit Invitrogen Pichia Fermentation Process Guidelines Video demonstration about Expression of Recombinant Proteins in the Methylotrophic Yeast Pichia pastoris in Hallam lab at UBC
| |||||||||||||||||||||