UCSF Chimera for 3D Printing
UCSF Chimera is a free and open source molecular visualizer. In this tutorial we’ll show how Chimera can be used to generate 3D printable molecular models for FDM/FFF desktop 3D printers. You can download Chimera at https://www.cgl.ucsf.edu/chimera/.
Finding PDB Files
First, you need the coordinate file of the molecule you want to print. The files containing the atoms coordinates for a molecule are called .pdb files (for protein data bank). These structures are determined using an array of techniques such as x-ray crystallography and nuclear magnetic resonance, and when they are published they are also deposited in databases and become freely available for download. The biggest such database is the Protein Data Bank at http://www.rcsb.org/
If you’re looking for cool structures and some explanations, the molecule of the month PDB articles at http://pdb101.rcsb.org/ are a good way to start browsing.
If you can't find your molecule, if it's not on the PDB it's probably because nobody knows it's structure yet. If what you want is some small molecule, you can try searching for the structure online, or can draw it using Avogadro, a free and open source molecule editor you can get at www.avogadro.cc.
Visualizing and Exporting Models
Once you have your PDB file, you can render it on a molecular visualizer. I’m using Chimera here, but the workflows is virtually the same if you use something else like PyMol or VMD.
When trying to visualize molecules, it is important to keep in mind there is no "right" image for them, we can represent atoms in a variety of manners that best present the features we want to convey. There are 4 different representations that are commonly used:
- Ribbons: depicts structural motifs (helices and sheets),
- Licorice or stick: show all the bonds between atoms
- Ball and stick: like stick, but also shows atoms as small spheres
- Spheres: the traditional representation most people know
- Surface: roughly the surface that another molecule (like water) would see. It’s obtaining by probing the spheres representation using a probe of arbitrary radius.
Guide for Chimera representations: https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/representation.html
For FDM 3D printing, surface representation is the easiest one to print successfully. If you want to print ribbons or lines, you must make sure that your model features are not too small, otherwise it will not print properly or will be too fragile. We’ll discuss several tips to address that, and once the ribbons have been thickened and extra bonds added, ribbon models will print well at an enlargement between 200% to 300%.
Once you have the representation that you want, you need to export an STL model by going to File > Export Scene, selecting STL as the file type, naming and saving your model.
In addition to the STL, it’s a good idea to save your Chimera Session by going to File > Save Session As. This is especially important because chimera has no undo button, and sometimes it takes several steps to revert an undesirable one click change.
A nice thing about Chimera is that it generates models at a consistent scale, at 10^7 magnification. This means that if you print at 100% scale, your model amplification is 10 million times. If you print at 300%, it’s 30 million times.
Finally, sometimes you want to use the Chimera command line to type commands instead of clicking around (often because there is no button for doing what you want). To access the Chimera command line, go to Favorites > Command Line, and it will appear in the bottom of the screen.
When making selections, you can save and name your selection by going to the menu Select > Name Selection, and then choosing a name for your selection. Then you can always access that selection using the command sel name, where name is the name of your selection.
Surface Rendering
Chimera standard STL rendering resolution is fine for most representations, but when printing surface models in large scale than the standard output, it's a good idea to bump up the amount of vertices to obtain a smoother model. The standard surface vertex density is 4, and a good amount is 10 if you want to enlarge your model by several times.
The easiest way to render the surface representation in high quality is to go to Actions > Surface > Show, or go to Presets > Interactive 3 (hydrophobicity surface). Then hit Presets > Publication (any of the 4 options work, the silhouettes and shadows don't affect STL quality). To do the same thing using command lines, just type surf to render the model surface. You can adjust the surface vertex density using the command setattr s density x (set attribute), where x is the density. For example, to set the density to 10, type setattr s density 10.0
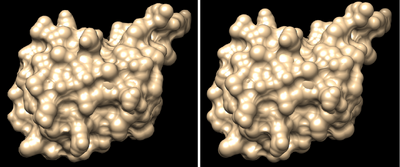
Chimera calculates a molecules surface by considering that each atom has a certain radius, and then seeing the depth that is accessible to a sphere of arbitrary radius. The default radius of that sphere is 1.5 angstrom, but you can change it using the command setattr s probeRadius x, where x is the new radius you want.
For example setattr s probeRadius 0.5 sets the probe radius to 0.5 angstrom. Below you can see examples of a surface rendered with the probe radius at 0.5 and 0.1. If you make the probe radius very small, you end up with the same shape as the spheres representation.
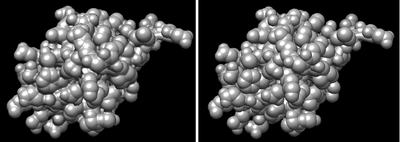
Sometimes your model is composed of various chains, and you just want to render the surface of one of them. One way you can do that is to use the split command, which separates the chains and generates a surface for each one.
For generating surfaces on arbitrary selections, use the command: split atoms selected
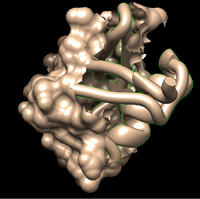
The other way is to delete the atoms of the sections you don’t want to render. If you merely hide them, your surface will appear broken, and not good for rendering a printable model.
Making Ribbons Fat
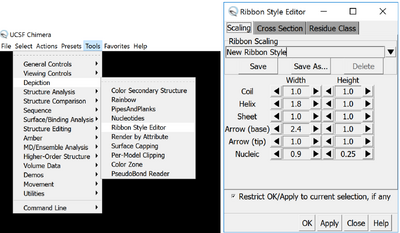
Change the ribbons appearance and make them as fat and thick as possible, by going to Tools > Depiction > Ribbon Style editor and changing the thicknesses. You want to make them as thick as possible, between 0.7 and 1. Keep an eye to make the sizes of the different elements match so that it looks good. You can also save your new ribbon style as a preset so you can readily reuse it.
Adding Hydrogen Bonds
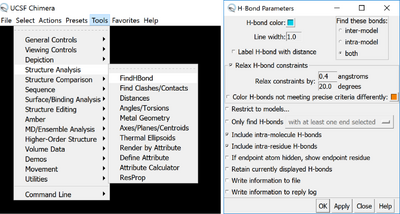
The other trick that to make the ribbons more stable is to show the hydrogen bonds, this makes beta sheets and alpha helixes super stable, and if the ribbon breaks, the whole model is less likely to fall apart. To do that, go to Tools > Structure Analysis > Find H bond, and you can make it show the H bonds.
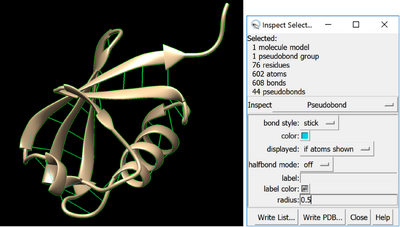
Then make them thick by selecting the whole model, going to Actions > Inspect. Select pseudobonds in the inspect option, and for bond style select stick.
Finally, you can make your bond thicker by altering the radius. The default is 0.2, and I’d suggest bumping it up to something about 0.5, so that they print fine and make the model sturdier, but are not too big.
It is important to keep in mind that H bonds will be shown for all atoms that are visible. So for example, if you make all atoms visible, including crystallographic water molecules, there will be a lot of spurious H bonds, and just hiding the water molecules will do away with most bonds that might be undesirable.
It is also important to note that if you render atoms as wires, they will not appear in your STL model, but a 3D H bond between them will appear. This can be useful if you’d like to show bonds between surfaces for example, but you don’t want to render the individual atoms. When rendering models with hydrogen bonds, it’s especially important to run the generate STL through Netfabb, because the STL mesh outputted by Chimera is not solid.
Adding Fake Bonds as Struts
You can add a fake bond between two atoms by adding a distance measure, and making it a cylinder. Go to Tools > Structure Analysis > Distances, and you can create a distance between any two atoms you have selected (you need to have only two atoms selected).
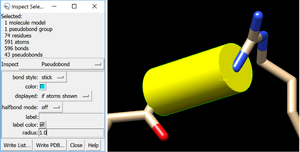
Then make them thick by selecting the whole model, going to Actions > Inspect. Select pseudobonds in the inspect option, and for bond style select stick. Finally, you can make your bond thicker by altering the radius. The default is 0.2, and I’d suggest bumping it up to something between 0.5 and 1, if you want this fake bond to hold the model together.
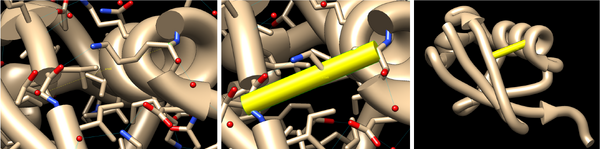
Such fake bonds can be added to the atoms of the protein backbone, so that their flat face don’t show. In order to see and select the backbone atoms, you must turn the ribbon representation off. After adding the fake bond, you can turn the ribbon on again, and hide the atoms, and the fake bond will look great.
Making Bonds Fat
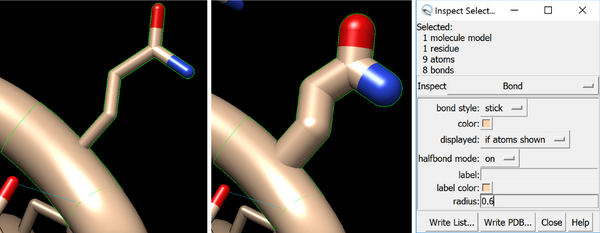
If you are using the licorice representation and want to print a group of atoms as part of a larger protein, such as a sidechain or a ligand, you can easily thicken them so that they will print better. Here I’ve selected a residue, which also selects the atoms from the sidechain. As with H bonds and distances (both of which are pseudobonds in chimera), you can change rela bonds thickness by going to Actions > Inspect. Select Inspect>Bond, and increase the radius.
Changing the Radius of Atoms
If you’d like to use the spheres representation, you can alter the size of individual atoms or bonds, or all the atoms of a given type. Just go to Actions > Inspect. Select Inspect Atom, and increase the radius. If you want to change the radius of all atoms of a given type, first select all of them by going to Select > Chemistry > Element, and choosing the atom type.
This can be very useful in certain cases. For example, consider the model of a triple base pairing below (which occurs in tRNA). In it the atom sizes were changed to be much smaller that the default. This highlights the spacing between the base pairs and make is easier to see how they are bonded. I also made each atom type a different size, so that even if printed on a single colour, it would be easy to recognize different atoms.
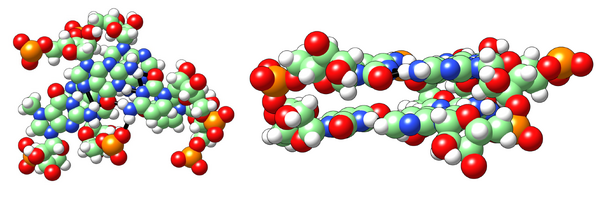
Chimera however has a huge problem if you use the spheres representation. When you export your STL, it will render each sphere with a huge number of faces, and if you have many atoms rendered as spheres your final file will be enormous. You can get around this problem by exporting to other 3D file format, such as X3D, and then converting that to STL. A way to perform this workaround is described here: http://www.cgl.ucsf.edu/pipermail/chimera-users/2014-June/010045.html Another way to get around that is to instead render your molecule as a surface, but make the probe radius very small with the command setattr s probeRadius 0.01. Chimera calculates a molecules surface by considering that each atom has a certain radius, and then seeing the depth that is accessible to a sphere of arbitrary radius. So if you make the probe radius very small, you end up with the same shape as the spheres representation. Don’t forget to increase the quality of the rendered surface using the command setattr s density 10.0, or even a quality higher than 10 if you want to enlarge your model a lot.
Mixing Different Representations to Obtain Great Models
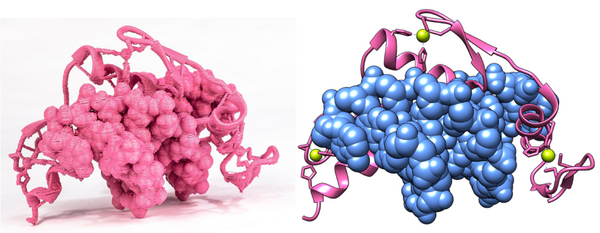
Using all the tips above, 3D printable models can be successfully prepared using any representation. This becomes especially powerful when different representations are mixed together to highlight different aspects of a model. My favourite example is the zinc finger model shown below. The DNA is rendered as spheres, the protein as ribbons, the residues interacting with the zinc atoms are shown as licorice, while the zinc atom itself is shown as a sphere. The hydrogen bonds and interactions of the zinc atoms have also been rendered, making the model sturdier.
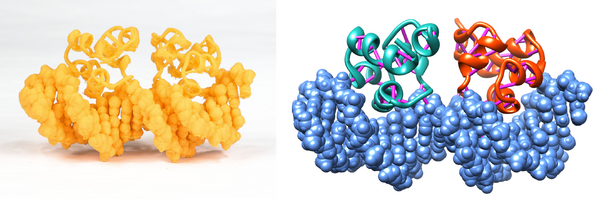
When working with models containing both protein and DNA, rendering the protein as ribbons and the DNA as spheres or surface is a straightforward way to highlight structural motifs and distinguish between protein and DNA. The cro434 model shown below is another example of this trick.
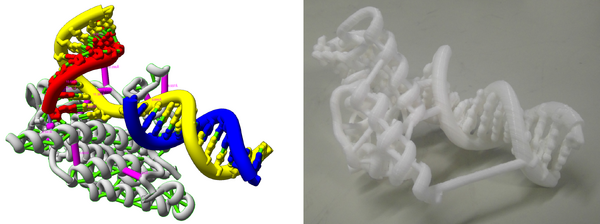
This is of course not the only possible way to do things - you have to be creative, and think through what you want to highlight in each model. For example, take a look at the model of RNA polymerase active site shown below. In it the nucleic acid backbone has been rendered as a thick coil, highlighting the strands conformation. The nucleic acids are shown as sticks, which allows us to understand how each piece of the strand if interacting and held together. A large chunk of protein is shown as ribbons, and the residues of the active site are also shown. A lot is going on. The entire model is made sturdy not only by rendering the hydrogen bonds, but also manually adding several thick fake bonds, shown in pink. Despite its complexity, this model printed very reliably, and is also sturdy.