Tiffany Guo
About me
Name: Tiffany Guo
Course/Minor: 20/5
Year of Graduation: 2009
Email: tiffguo
Current Classes: 5.03, 6.00, 20.109, 21F.108
Module 1: Genome Engineering
Day 1: Start up Genome Engineering
| protein | function | re-engineering ideas |
|---|---|---|
| I | assembly | modify gene to that multiple channels can be made, and thus mature phage secretion is accelerated |
| II | replication of DNA + strand | modify so it can nick foreign DNA (e.g. the E. Coli's) and the phage can replicate and package a portion of the host DNA |
| III | phage tail protein (5 copies) | modify end of protein so that it can bind to other cells (and infect other cells) besides E. Coli |
| IV | assembly | modify gene to that multiple channels can be made, and thus mature phage secretion is accelerated |
| V | binds ssDNA | allow it to sequester double stranded DNA also, then it can be used as a vector for infecting cells with desired DNA fragments. |
| VI | phage tail protein (5 copies) | delete to learn more about its function. |
| VII | phage head protein (5 copies) | delete to learn more about its function. |
| VIII | phage coat protein (2700 copies) | tag it (perhaps with flourescence) to learn more about phage coat assembly |
| IX | phage head protein (5 copies) | modify so that it can act similarly to pIII |
| X | DNA replication | make pX more active so that more + strands will accumulate, allowing the host cell to produce even more phages. |
| XI | assembly | modify gene to that multiple channels can be made, and thus mature phage secretion is accelerated |
Day 4: Genome Refactoring
I refactored the phate genome to remove overlaps in the genome and to add in restriction enzyme sites between each part. I edited codons to retain amino acid sequence while weakening the rbs or promoter. Considerations: 1) eliminate rbs and promoters within genes: took into consideration codon changes that would not change amino acid sequences but would decrease the strength of the rbs or promoter. 2) add restriction sites between each part: tried to keep sticky ends as consistent as possible (i.e. after every promoter, the restriction site sticky end is 3'ctag), however, there aren't enough zero cutting isoschizomers
On the parts registry, my part is BBa_M31975.
New Parts: a) restriction enzyme sites
| restriction enzyme | code | parts number |
|---|---|---|
| Acc65I | G^GTAC_C | BBa_M31997 |
| AvrII | C^CTAG_G | BBa_M31994 |
| AscI | GG^CGCG_CC | BBa_M31990 |
| BsiWI | C^GTAC_G | BBa_M31996 |
| SpeI | A^CTAG_T | BBa_M31992 |
| BssHII | G^CGCG_C | BBa_M31989 |
| BsrGI | T^GTAC_A | BBa_M31995 |
| MluI | A^CGCG_T | BBa_M31988 |
| BclI | T^GATC_A | BBa_M31987 |
| EagI | C^GGCC_G | BBa_M31986 |
| BglII | A^GATC_T | BBa_M1985 |
| NheI | G^CTAG_C | BBa_M31993 |
| PspOMI | G^GGCC_C | BBa_M31984 |
| EcoRI | G^AATT_C | BBa_M31983 |
| XbaI | T^CTAG_A | BBa_M31991 |
| NotI | GC^GGCC_GC | BBa_M31982 |
| MfeI | C^AATT_G | BBa_M31976 |
b) Trancription terminator
| part number | code |
|---|---|
| BBa_M31977 | tacaattaaaggctccttttggagcctttttttt |
c) coding sequences:
| gene | parts number | parts removed | # bp changed | code (bp changed are capitalized) |
|---|---|---|---|---|
| 5 | BBa_M31981 | rbs 7, restriction site BsrGI | 3 | atgattaaagttgaaattaaaccatctcaagcccaatttactactcgttctggtgtttctcgtcagggcaagccttattc
actgaatgagcagctttgttacgttgatttgggtaatgaatatccggttcttgtcaagattactcttgatgaaggtcagc cagcctatgcgcctggtctgtaTaccgttcatctgtcctctttcaaagttggtcagttcggttcccttatgattgaccgt ctgcgcctcgttccAgctaaCtaa |
| 7 | BBa_M31980 | rbs 9, promoter 8 | 6 | atggagcaggtcgcggatttcgacacaatttatcaggcgatgatacaaatctccgtAgtactGtgtttcgcgctGggCat
TatcgctgggggCcaaagatga |
| 9 | BBa_M31978 | rbs 8 | 1 | atgagtgttttagtgtattctttcgcctctttcgttttaggttggtgccttcgtagtggcattacgtattttacccgttt
aatggaaacttcAtcatga |
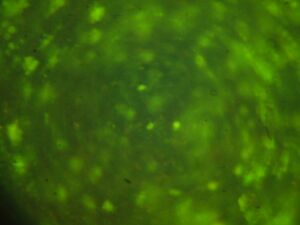
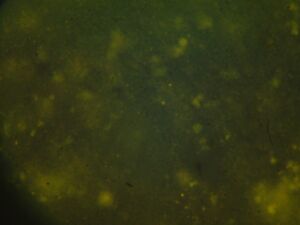
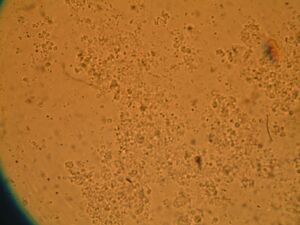
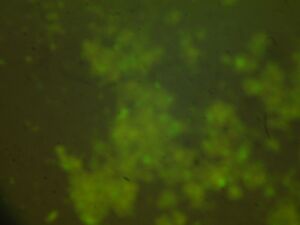
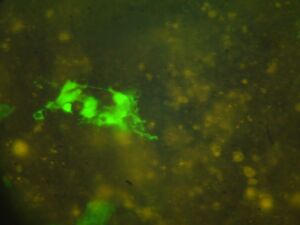
Module 2: Calcium Signalling
Day 4: Calcium Signalling In-Vivo
| DNA | Mock | 1/2 DNA | |
|---|---|---|---|
| Mock |   |
  |
  |
| Untreated |   |
  |
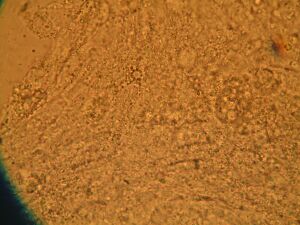 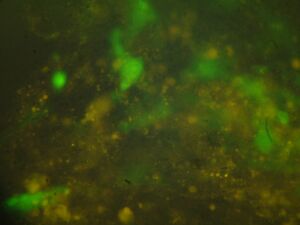 |
| Histamine (0.2mM) | 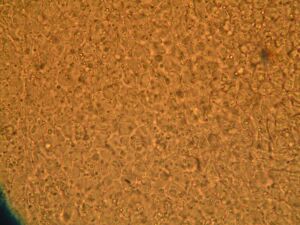 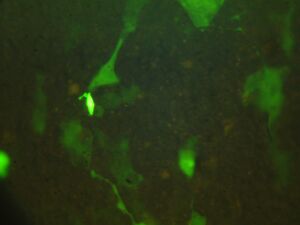 |
  |
  |
| EGTA (2mM) |  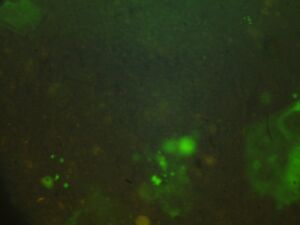 |
  |
  |