The Microbiome
Definition
The microbiome is most commonly thought of as the collection of the estimated 1014 microbes, the microbiota, that reside in the gut (1). The term was coined by Joshua Lederberg in 2001, yet many people are undecided about the exact meaning of the word (2). In general, microbiome can refer to any genetic collection of microbes in a specific region, but here it will be used synonymously with the microbiome mentioned previously. The microbiome plays a role in digestion and nutrient uptake, metabolism, and the immune system among many other things. It is easily modified by diet, infection, and many other outside factors, and is thought to be linked to various immune disorders.
Background
The microbiome is always changing, and is initially inherited at birth from the mother. The development of the microbiome is something that is still under questioning. In one study, an infant male was sampled from birth for 2.5 years and the development of his microbiome was documented throughout (3). This one study showed that the infant microbiome composition, previously described as chaotic, is likely dependent on life events such as the switch from breastfeeding to solid food, or the introduction of antibiotics to combat one of the many common infant illnesses. Diversity of the microbiome increases over time from birth through infancy, though the diversity is typically affected positively by the introduction of new foods, and negatively by the use of antibiotics or by illness.
Factors Affecting the Microbiome
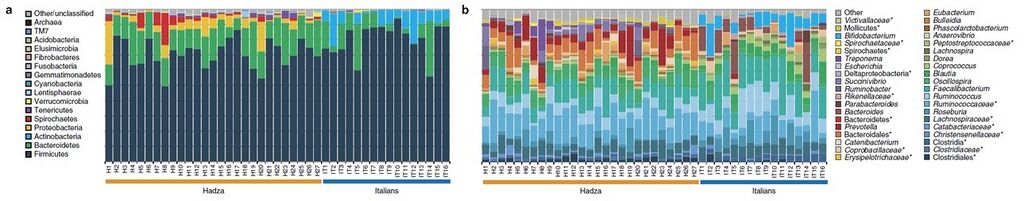
As of 2014, studies are being performed questioning the limits and variability of the human microbiome due to changes in the many factors that can affect it. One group studied the variability of the microbiome, or lack thereof, in 16 Italian urban citizens, rural African farming groups, and 27 members of the Hadza of Tanzania – a group of hunter-gatherers (4). The Hadza and Italian citizens both had a mean age of 32 years. Testing was done using voluntarily provided fecal samples to gather information about the microbiota. In the Hadza, they found unique differences in the composition of the microbiome that may have resulted from the male-hunter, female-gatherer lifestyles that they lead. Also, they found a lack of biodiversity in the urban Italian participants when compared with the Hadza, shown in Figure 1 (4). This compliments the trend found by other studies between urban and rural societies, all due mainly to dietary differences (5). However, diet is not the only factor that kills biodiversity in the microbiome of the urban populace. Antibiotics and other medicines, used heavily in urban society, as well as pollutants and obesity all contribute to this decline in biodiversity of the microbiome (6).
Further studies have been carried out using mice to investigate the differences in the microbiome between genders. Hormonal differences have shown to create differences in the immune system, specifically involving diabetes (7). It was observed in nonobese diabetic mice that type 1 diabetes developed in twice as many females as males in specific pathogen free conditions, due to the lack of testosterone. In germ free conditions, there was no definitive trend between the sexes, so the development of type 1 diabetes is assumed to also depend on commensal microbes – those on the exterior of the body - as well as on
Relevance in Disease
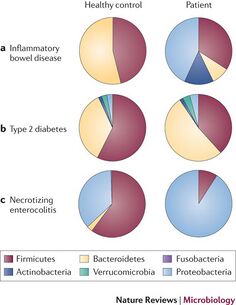
Large scale changes in diet and living conditions over time can cause changes to the microbiome, as discussed earlier. However, there is evidence that disturbing the microbiome, called dysbiosis, can result in diseases of the intestines and surrounding organs (9). An example of the correlations between disease and the microbiome can be seen in Figure 2. Inflammatory disease can be triggered by both pathogenic microbes and other microbes that would normally be controlled by a stable state of the microbiome and by proper immune response. It has been theorized that there are only a limited amount of stable states of the microbiome, the rest possibly resulting in dysbiosis.
Inflammatory Bowel Diseases
The diversity of the microbiome is closely associated with multiple inflammatory bowel diseases (10). This was shown using 16S rDNA based analysis in one study. Fecal samples were taken from patients with either ulcerative colitis and Crohn's disease, both active and inactive. These samples were then analyzed and compared with healthy control subjects. The diversity of the microbiome in ulcerative colitis was reduced to 30% in a 31 patient sample, and Crohn’s disease (CD) reduced to 50% in a 26 patient sample, when compared with 46 controls. This was concluded to be due to loss of certain species of bacteria in the microbiome, which was verified using polymerase chain reaction. In contrast to this reduction in diversity found by one group, another group instead observed an unchanging microbe diversity in the microbiome, rather the presence of different species of bacteria in patients with CD (11). This second study also used 16S rDNA analysis. It was observed that Bacteroides and bifidobacteria were less represented in patients with CD than in control subjects without the disease, but enterobacteria was detected in all CD patients and not in control subjects.
Obesity
Obesity has been associated with an estimated 50% reduction in Bacteroidetes, and a large portion of Firmicutes in studies done using mice (12). The microbiome of a mouse has not been proven completely similar to that of a human, but the Bacteroidetes and Firmicutes are the most abundant bacteria found in mice and in humans, making this finding rather significant. When a mouse is subject to an increase in “obese microbiota”, the total body fat of the mouse increases significantly over a mouse that is subject to “lean microbiota” (13). These terms are used to loosely represent the composition of the microbiome of an obese mouse vs. the microbiome composition of a lean mouse. This was done by extracting DNA from obese and lean mice by phenol-chloroform extraction, purifying it, and injecting it into test subjects.
Liver Cancer
One study showed that obesity promotes liver cancer by altering metabolite amounts in the microbiome (14). This leads to increases in deoxycholic acid which provokes the release of senescence-associated secretory phenotype (SASP). SASP has been shown to cause cancer in trials performed on mice. The possibility that hepatocellular carcinoma development, a human liver cancer, is tied to intestinal bacteria was also studied. This was done by treating mice with antibiotics, to reduce the bacteria in the intestine, and observing a reduction in hepatocellular carcinoma. However, there were some inconsistencies in mice lacking a certain gene, which left this theory unresolved. As a result of this work, high concentrations of deoxycholic acid, caused by dysbiosis due to obesity, were shown to contribute to liver cancers in mice. Furthermore, this conclusion was tested using human hepatic stellate cells, and the results were consistent.
References
1. Honda, K.; Littman, D. The Microbiome in Infectious Disease and Inflammation. Annual review of immunology 2012, 30, 759-795.
2. Lederberg, J.; McCray, A. 'Ome Sweet 'Omics - A Genealogical Treasury of Words. Scientist 2001, 15(7) (8).
3. Koenig, J. E.; Spore, A.; Scalfone, N.; Fricker, A. D.; Stombaugh, J.; Knight, R.; Angenent, L. T.; Ley, R. E. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America 2011, 108, 4578-4585.
4. Schnorr, S. L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; Fiori, J.; Gotti, R.; De Bellis, G.; Luiselli, D.; Brigidi, P.; Mabulla, A.; Marlowe, F.; Henry, A. G.; Crittenden, A. N. Gut microbiome of the Hadza hunter-gatherers. Nature Communications 2014, 5.
5. De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J. B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (33), 14691-14696.
6. Ehlers, S.; Kaufmann, S. H. E.; the Participants of the 99th Dahlem Conference. Infection, inflammation, and chronic diseases: consequences of a modern lifestyle. Trends in Immunology 2010, 31 (5), 184-190.
7. Markle, J. G. M.; Frank, D. N.; Mortin-Toth, S.; Robertson, C. E.; Feazel, L. M.; Rolle-Kampczyk, U.; Bergen, M. v.; McCoy, K. D.; Macpherson, A. J.; Danska, J. S. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 2013, 339, 1084-1088.
8. Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews Microbiology 2011, 9, 279-290.
9. Littman, D. R.; Pamer, E. G. Role of the commensal microbiota in normal and pathogeni host immune responses. Cell Host Microbe 2011, 10, 311-323.
10. Ott, S. J.; Musfeldt, M.; Wenderoth, D. F.; Hampe, J.; Brandt, O.; Folsch, U. R.; Timmis, K. N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685-693.
11. Seksik, P.; Rigottier-Gois, L.; Gramet, G.; Sutren, M.; Pochart, P.; Marteau, P.; Jian, R.; Dore, J. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 2003, 52, 237-242.
12. Ley, R. E.; Backhed, F.; Turnbaugh, P.; Lozupone, C. A.; Knight, R. D.; Gordon, J. I. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America 2005, 102, 11070-11075.
13. Turnbaugh, P. J.; Ley, R. E.; Mahowaldl, M. A.; Magrini, V.; Mardis, E. R.; Gordon, J. I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027-1031.
14. Yoshimoto, S.; Loo, T. M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; Honda, K.; Ishikawa, Y.; Hara, E.; Ohtani, N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97-101.