The BioBricks Foundation:BBFRFC15
Purpose
BioBrick standard assembly or BBa places limitations on the sequence content of parts, controls the order in which BioBrick parts can be combined, and introduces sequence artifacts into BioBrick part assemblies. Since these limitations make it hard to create protein fusions, make screening libraries, and domesiticate some parts to the BioBrick standard, there is a need to create a new, upwardly compatible BioBricks assembly standard or BBΩ that will allow the creation of any DNA sequence in any order. By introducing BioScaffold parts, which are a new set of BioBrick compatible parts, standard assembly can become upwardly compatible. This will allow standard assembly to meet the needs of the Synthetic Biology community while at the same time retaining the advantages of the large registry of existing BioBrick(R) standard biological parts. The community is invited to assist in the development, fabrication, testing, and application of BioScaffold parts. This document is intended to be static and thus the definitive version will appear on the document's publication date. However, the community is invited to comment on this draft standard either formally with a new BBFRFC or informally on the BBFRFC15 comments page [1]. The community is also invited to assist in the development, fabrication, testing, and application of BioScaffold parts. Community members can view current progress at the BioScaffold Parts Page [2] and state their commitments to advance the development of BioScaffold parts at the BioScaffold Parts Users' Group Wiki [3].
Abstract
This document describes a new family of BioScaffold parts that have the potential to extend the BioBrick standard to allow the user to create almost any part or subpart. BioScaffold parts are temporary parts that can easily be added or removed from BioBrick assemblies, facilitating their eventual replacement with permanent BioMortar parts for the creation of protein fusions, library constructions, or part domestication. The original goals of standard assembly were to create interchangeable DNA parts, to assemble the sub-parts needed to create a whole part, to make DNA fabrication a commodity, and to gain the ability to use parts or sub-parts created by others. The current limitation in standard assembly is that it does not allow users to create all the types of parts and subparts or manipulate the parts in the order that they desire. Specifically, BioBrick standard assembly specifies the positions at which new parts can be added (to either end of an existing part), creates artifactual DNA scars between two DNA parts when they are composed, and specifies that certain restriction enzyme recognition sites can only occur in the prefix and suffix regions of parts (EcoRI, XbaI,SpeI, and PstI.) New standards have been proposed to overcome these limitations, yet most greatly reduce interchangeability of parts or place strong restrictions on the types of parts the user can create. BioScaffold parts allow high level DNA manipulations to occur independent of the standard assembly part composition process AND still maintain the standard BioBrick format of the final assemblies that result. The new family of parts has the potential to enable a variety of innovations in part composition to be implemented without changing the format of the BioBrick parts or the use of standard assembly to compose the parts. In the proposed usage, BioBrick and BioScaffold parts will first be composed using standard assembly, second temporary BioScaffold parts will be removed and replaced by permanent BioMortar parts or other BioScaffold parts, and finally the desired final BioBrick assembly will be created.
Key Words and Phrases: standard assembly, BioBrick(R) standard biological parts, gene synthesis, part composition, protein fusions, library construction, part domestication or integration, error correction, and rapid prototyping
Related Requests for Comments: BBFRFC9 [4], BBFRFC10 [5], BBFRFC12 [6], BBFRFC13 [7], BBFRFC14 [8], and potentially BBFRFC3 (not yet posted)
Historical Overview and Introduction to BioScaffold Parts
Motivation
As BioBrick(R) standard biological parts have found application within the Synthetic Biology community, their limitations and the limitations of the standard assembly process continue to hinder the rapid incorporation of new developents. One emerging approach to overcome these limitations is to scrap BioBrick standard assembly [1,2] and replace it with a set of customized assembly standards to address specific situations (as is currently occurring for protein fusions [3,4,5,6,7,8, 9,10] and parts libraries [11]) though this strategy has the potential to dilute the utility and value of BioBricks(R) standard biological parts since it incentivizes the creation of new divergent standards rather than convergence of both parts and standards. Divergent standards evolution has troubling consequences for the future of BioBrick(R) standard biological parts and the Registry of Standard Biological Parts, as the best parts of a certain class may either be effectively taken out of circulation if they do not meet the current, most fashionable standard or if the credit for a part’s creation is directed simply to the person who converts it to the most fashionable standard. Such results are deleterious because they have the potential to slow progress in the field. Rather than support the evolution of divergent standards, we instead propose to create an assembly standard BBΩ and a new family of BioBrick (R) standard biological parts that are specifically designed to overcome the known limitations of standard assembly and which favor evolution of parts of increasing quality resulting in a final static representations within the Registry of Standard Biological Parts.
Limitations of Standard Assembly
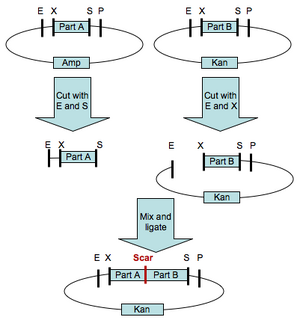
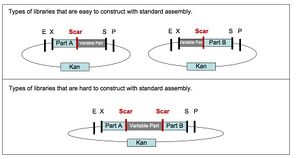
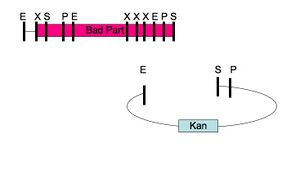
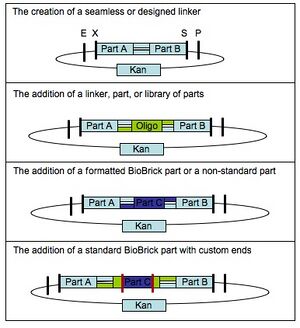
Any two DNA sequences that are BioBrick(R) standard biological parts can be easily composed to form a larger BioBrick(R) part using standard assembly [10,11]. However, standard assembly has several limitations. First, the assembly process introduces a short DNA scar between the sequences of the two original parts (see Figure 1.) The scar has the potential to be deleterious for certain applications, such as protein engineering [3,4], where two protein parts should ideally be connected either seamlessly or with an intermediate linker that is not constrained by scar sequence. Second, standard assembly allows composition of only two parts at a time (see Figure 1.) For library construction the ability to compose only two parts at a time is a key disadvantage, since in order to maintain library complexity the variable portions of a library assembly must be added last (see Figure 2.) Third, standard assembly uses four restriction enzymes, which must be removed before the part can be used as a BioBrick(R) standard biological part (see Figure 3.) For some sequences, removal of all the restriction enzymes is a time consuming process and accepting new standards reliant on additional restriction sites places additional burdens on part creators. Fourth, standard assembly may have additional limitations or shortcomings that will be revealed as BioBrick(R) parts continue to find use in new applications and new or competing technologies develop.
The Synthetic Biology Community's Improvements Upon Standard Assembly
The Synthetic Biology community has recognized the flaws of standard assembly and has proposed many alternative standards for parts composition that both build upon standard assembly and upon each other. The proposed standards each improve upon standard assembly in a unique way, yet no one standard incorporates the advantages of the others. At this point selecting any one standard means forgoing the benefits of many of the others, placing the field in a dilemma which we hope to resolve with the introduction of the BioScaffold parts. The following groups have made valuable contributions and improvements to standard assembly. Austin Che [3] and Pamela Silver [4] determined that the original BioBrick (BBa) parts and standard assembly were not conducive to the construction of protein fusions and developed the first proposed alternatives. Chris Anderson [9] developed a new assembly standard where the scar created from the composition of two parts contains the DNA code for the classic “glycine-serine” protein linker. Katja Arndt and the Freiburg iGem2007 team [9,13] found a way to make protein fusions while allowing the parts to retain the format of the original BioBricks standard. We define the ability to improve upon standard assembly while retaining the ability of parts to be composed by standard assembly as "The Freiburg Convention." Raik Gruenberg [9] developed a way to convert between parts from different proposed standards, in this case from the Freiburg format to the Silver format. Knight [5] recognized the need to use heat killable restriction enzymes in assembly schemes in order to retain the ability to perform automated assembly. Austin Che [3] pioneered the use of offset cutters in a proposed assembly scheme and saw the potential for ligation independent cloning using offset cutters. Sergio Peisajovich, Wendell Lim, and the UCSF 2007 iGEM team [10] showed the potential of using offset cutters for multipart ligations which has the potential to evolve into a method for enabling rapid prototyping of parts. New innovations are also developing in the area of library creation, which may become a critical technology and tool for developing functional BioBrick devices. Chris Anderson [11] and Zhan Jian of the 2007 USTC iGEM team [11] demonstrated a way of making vectors to help screen libraries of parts.
The Intrinsic Conflict Between Part and Device Designers
If BioBrick(R) standard biological parts and standard assembly are retained in their present form, there will always be tension between parts designers who wish to break all the rules of standard assembly and the device designers who would like to use simple, reliable methods of parts composition so that they can focus on challenges at different levels of abstraction. Standard assembly and the proposed new standards currently favor the needs of the device designer. However, little development has been made in the direction of creating tools that parts designers to break the rules of standard assembly in a standard fashion and at multiple levels of hierarchy. If tools can be created that allow parts designers to effectively break the rules of standard assembly, enable use of standard assembly when it is desirable, and facilitate restoration of parts to BioBrick from before they are handed off to parts designers, then this tension between parts designers and parts creators can be resolved without fragmentation of standard assembly into many incompatible domain specific standards.
Functional Description of BioScaffold and BioMortar Parts
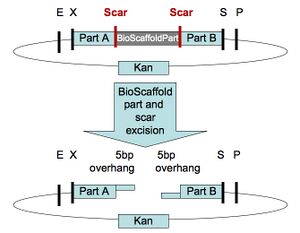
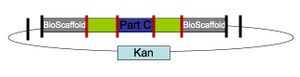
In standard assembly one BioBrick part is added to another BioBrick part on the right or left side, creating a new BioBrick part with a scar in the middle (Figure 1.) BioScaffold parts can be added to the left or right side of BioBrick parts in the same manner as a BioBrick part. When one is done creating the BioBrick assembly, one can cut out the BioScaffold part from an assembly by adding either one or a few enzymes and purifying the resulting product with either gel purification or a spin column (Figure 4.) After the BioScaffold part is cut out a few options for what to do next are available (Figure 5.) Usually the options include adding a new part where the BioScaffold part used to be, since cutting out the BioScaffold part leaves behind sticky DNA ends that allow addition of a new DNA part (Figure 4.) If the ends can fuse back together without adding a new part, then the operation is the equivalent of adding a null part or a null BioMortar part to the assembly (Figure 5,) where BioMortar parts are the DNA additions that can be used to replace temporary BioScaffold parts. BioScaffold parts can also be replaced with a BioMortar parts that are made out of annealed DNA oligonucleotides (Figure 5.) BioScaffold parts can also be replaced DNA that is fabricated by different methods. PCR can be used to create a BioMortar part, but requires introduction of appropriate restriction enzyme sites at each end of the sequence. The restriction enzyme sites can then be cut with an enzyme to create the new BioMortar part (that has appropriate ends that will allow it to replace the BioScaffold part) (Figure 5.) In addition, oligonucleotides and BioScaffold parts can be incorporated into BioBrick assemblies to help create custom BioMortar parts in a different fashion (Figure 6.)
BioScaffold Parts Definitions
BioBrick Prefix, Suffix, and Scar BioScaffold parts BioBrick(TM) standard biological parts that contain characteristics that enable their full or partial removal from BioBrick constructs or assemblies.
BioMortar parts Parts that replace BioScaffold parts after their full or partial removal from a BioBrick construct or assembly. Thus, BioMortar parts do not need to be composed by standard assembly or subject to the BioBrick standard to be integrated into a BioBrick construct or assembly as a BioMortar part.
Classes of BioScaffold parts Class α, β, γ, and mixed BioScaffold parts refer to parts that perform as described below when placed within an assembly of the form <Scar><BioScaffold Part><Scar>. BioScaffold Class α (alpha) parts outermost cuts occur outside of scars surrounding BioScaffold part BioScaffold Class β (beta) parts outermost cuts occur within scars surrounding BioScaffold part BioScaffold Class γ (gamma) parts only cut within the BioScaffold part BioScaffold Class δ (delta) parts and beyond will be defined as the warranted by the community Mixed parts Contain properties of several classes. Class γ parts cannot form mixed parts at this time given the current definitions of BioScaffold part classes.
BioScaffold external enzymes. Enzymes with the outermost recognition sites that cut within or outside the scar when a BioScaffold part is placed within an assembly of the form <Scar> <BioScaffold Part> <Scar> or into the prefix or suffix region when a BioScaffold part retains its original form <Prefix><BioScaffold Part><Suffix>(i.e., these enzymes do not cut solely within the BioScaffold part though they may cut partially into it.) Enzymes with this function tend to have long (six base pair or longer) recognition sites in order to decrease their frequency. They may also have special properties such as a recognition site that is far from the cut site (for example, PsrI and PpiI.)
BioScaffold internal enzyme Enzymes that only cut within the BioScaffold part. Usually these are inserted to facilitate removal of a BioScaffold part using Qiagen spin columns (PCR cleanup kit) or to prevent religation if the BioScaffold part is improperly excised from an assembly. Enzymes with this function tend to have long (six base pair or longer) recognition sites in order to decrease their frequency. They may also have special properties such as cut probabilities that vary as a function of GC content (for example, AscI and PacI) or enable designs that prevent religation after cutting (for example MabI.)
Overhang or sticky end parameters Strand, length, and compatibility of DNA overhangs created by cutting with the outermost (or most relevant) BioScaffold external (or internal) enzymes.
The current class of BioScaffold parts (for example the current family of BioScaffold α parts, consisting of BBa_J70010, BBa_J70012, BBa_J70030, and the composite part BBa_J70032) are subject to several additional design considerations beyond their ability to enable their removal from BioBrick assemblies. The current class of BioScaffold parts contain internal recognition sites for the BioScaffold specific enzymes PpiI and/or PsrI. BioScaffold specific enzymes are enzymes that enable the BioScaffold part to be removed from an assembly when the appropriate enzyme is added. For the BioScaffold α class of parts PpiI and PsrI were selected as the specific part removal enzymes because they have rare recognition sequences (which are seven base pairs long and have additional positional constraints), they have reasonable cutting efficiencies (though PpiI is more efficient), they are capable of being heat killed (allowing them to be used in automated assembly strategies), and due to their recognition sequences can be utilized to allow excision of BioScaffold parts to occur outside the adjoining BioBrick scars. Within the BioScaffold α class parts, PsrI and PpiI sites have been arranged such that the characteristic five base pair ends are formed upon excision of the BioScaffold parts (as shown in Figure X. This would in theory allow the adjoining BioBrick parts to be ligated if their sequences are compatible as shown in Figure Y.) The BioScaffold α class of parts also contain internal restriction enzyme recognition sites. For the current parts the internal restriction enzymes include MabI and (AscI or PacI). These enzymes can be used in some situations to help facilitate the removal of BioScaffold parts using Qiagen PCR cleanup spin columns (which help remove double stranded DNA of up to 45 base pairs in length) and thus help to prevent reannealing of a part if incomplete BioScaffold part removal occurs. It is optional which of these enzymes to use, though PacI sites are helpful if the surrounding parts have a high GC content and AscI is useful if the surrounding parts have a low GC content. The current class of BioScaffold α parts can be expanded by changing the internal restriction enzyme sites present.
The Protein Fusion Problem
BioBrick parts and standard assembly do not currently facilitate protein fusions. Since protein fusions are currently a small fraction of the parts synthetic biologists as a whole make, one approach is to simply decree that standard assembly does not support protein fusions. However, the community has developed new standards for making protein fusions, showing that they highly value the benefits of standard assembly. Now that these proposed standards have been created, a natural approach is to pick the best of these standards and convert every existing part over to it. However, protein fusions are unlikely to be the one construct that standard assembly and BioBrick parts do not support. Standard assembly needs to be made more flexible so that it can support a variety of improvements.
BioBricks and standard assembly do not allow the creation of protein fusions, as their composition prevents translation to continue directly from one protein part to another. The protein fusion standards proposed by the community allow one to compose two protein parts in a standardized fashion, but first require conversion or domestication of each part to the chosen standard (conversion to a standard is not always simple, even when converting a native part to a BioBrick part.) The family set of BioBrick parts introduced here, the BioScaffold parts, allow creation of protein fusions in many forms. They allow many protein parts in the registry to be directly translated into protein fusions in a customized fashion, with a standard methodology. At the same time, they incentivize standard forms for proteins that frequently fused with others such as protein tags. Thus, they satisfy the needs of both parts and device designers.
A Protein Fusion Strategy for Parts Designers: Using Off the Shelf Parts and Custom Linkers
Parts designers would like to take any two parts from the registry and connect them as they see fit. BioScaffold parts can enable this type of operation and are especially well suited to the construction of protein fusions by this method.
In order for protein parts to be compatible with this method, three requirements are placed on the parts. The protein parts should ideally begin with G (a BioBrick Prefix containing AT should precede them, completing the start codon) and end with TAA (instead of TAA TAA for instance.) See [ref] and appendix for a description of the BioBrick Prefix. Also the protein parts should not contain the PpiI restriction enzyme which is currently needed for removing protein tails. If the part contains a PpiI restriction enzyme site but not a PsrI restriction enzyme site, then the user should think about converting the part into a protein fusion part capable of automated assembly (see section below.) PpiI and PsrI have seven base pair recognition sites and thus are fairly rare. If the part has an unusual start (ATG) or end codon (TAA), then the user has three choices: see if can still work in the assembly, convert it to an appropriate protein fusion capable part (either for parts or device designers), or find a way to use it without modifying it using a combination of BioScaffold or BioMortar parts.
Example: Constructing a Protein Fusion Using BioBrick Parts and a Custom Linker
In order to place a custom linker between two BioBrick parts, first place a BioScaffold part between two BioBrick parts using standard assembly. (If you are using two protein parts and want to place a flexible linker between them you will need to use a BioScaffold part that removes protein tails, for instance BBa_J70030, or a part that removes both protein heads and protein tails, such as BBa_J70032. The vectors for the assembly process should be chosen to meet the requirements for 3A assembly.) See [ref] and appendix for DNA level descriptions of the BioBrick "Prefix," "Scar," and "Suffix" described below.
Table 2: Placing a BioScaffold part between two BioBrick parts First compose the prefix protein and BBa_J70030 using 3A assembly: <EcoRI cut Prefix w/AT>G protein TAA<SpeI cut Suffix> + <XbaI cut Prefix w/AG> BBa_J70030 <PstI cut Suffix>+<EcoRI and PstI cut Vector> Next compose the suffix protein with the composite prefix protein and BBa_J70030 part: <Prefix w/AT>G protein TAA<Scar w/AG> BBa_J70030 <SpeI cut Suffix>+ <XbaI cut Prefix w/AT>G protein TAA<SpeI cut Suffix>+<EcoRI and PstI cut Vector> Now you have a BioScaffold part composed between two BioBrick (protein) parts: <Prefix w/AT>G protein TAA <Scar w/AG> BBa_J70030 <Scar w/AT>G protein TAA<Suffix>
Now you will need to completely cut out and remove the BioScaffold part. Note that when the BioScaffold part is cut out, some adjoining sequence can be removed. (For example when using BBa_J70030, the protein tail and part of the protein head is removed.)
Table 3: Removing the BioScaffold Part
First you should cut with the BioScaffold part specific enzyme. (For BBa_J70030 it is PpiI.)
Location of PpiI cut sites in the assembly:
<Prefix w/AT>G . . . ppppp^TAA<Scar w/AG> BBa_J70030 <Scar w/AT> Gssss^. . . TAA<Suffix>
^ppppp ^Cssss^
This leaves BioScaffold part fragments that are too large to be removed with a Qiagen
PCR cleanup spin column. So first cleanup using the spin column and then cut with an
intermediate BioScaffold part enzyme. (For BBa_J70030 it is MabI or PacI.)
Then cleanup with a Qiagen PCR cleanup spin column to completely remove the BioScaffold part:
<Prefix w/AT>G . . . ppppp^ ^. . . TAA<Suffix>
^ ^Cssss
Now design a custom oligonucliotide part that will link together the two separate proteins. Usually the linker should code for a flexible linker (containing glycines and serines.) The main requirements here are to have an annealed oligonucleotide melting temperature greater than 30 degrees Celsius, so that the part can be ligated into the assembly, and a length than can be synthesized at reasonable cost (usually less than 60 base pairs.) It is also necessary to make sure that the protein translation will occur in frame after the linker.
Table 4: Creating a BioMortar flexible linker part to replace the BioScaffold part
Glycine-Serine-Glycine-Serine linker with a melting temperature of 38 degrees Celsius.
5'-GGT TCC GGG TC Gssss-3'
3'-ppppp CCA AGG CCC AG-5'
Gly-Ser-Gly-Ser
The oligo can now be annealed and ligated between the protein parts (restoring them to their orginal form with a flexible linker joining them.)
Table 5: The final construct
<Prefix w/AT>G protein GGT TCC GGG TCG protein TAA<Suffix>
CCA AGG CCC AGC
Gly-Ser-Gly-Ser
A Protein Fusion Strategy for Device Designers: Automated Assembly and Defined BioBrickPF Parts
Protein designers do not want to have to design a custom part every time they wish to make a protein fusion. They want to be able to take protein parts off the shelf and be able to immediately put them together into protein fusions. This may mean that they will work with different types of parts than protein designers. For instance they may place a premium on well tested parts that have been used in creating protein fusions before when designing their system. Ideally, creating a part that can be used by device designers should be the last step in the design process by a part designer. When a part is handed off in this form, the device designer will gain an additional measure of confidence in using the part in the future.
Format of BioBrickPF parts
Prefix Protein Fusion Preparation Part (BioBrickPF-P): Composition of a BioBrick protein part and a BioScaffold part that can be cut with the BioScaffold part specific enzyme, leaving characteristic ends.
Table 6: Format of the BioBrickPF-P (protein and BioScaffold) composite parts BioBrickPF-P(Type 1) Part:(Prefix Protein has a tail, BioScaffold specific enzyme is PpiI) <Prefix w/AT>G protein GGATCC TAA <Scar w/AG> BBa_J70030 (PpiI) <Suffix> BioBrickPF-P(Type 2) Part:(Prefix Protein lacks a tail, BioScaffold specific enzyme is PpiI) <Prefix w/AT>G protein GGATCC <Scar w/AG> BBa_J70010 (PpiI) <Suffix> BioBrickPF-P(Type 3) Part:(Prefix Protein lacks a tail, BioScaffold specific enzyme is PsrI) <Prefix w/AT>G protein GGATCC <Scar w/AG> BBa_J70012 (PsrI) <Suffix>
Table 7: Format of the BioBrickPF-P parts after excision with the BioScaffold specific enzyme:
<Prefix w/AT>G protein G GATCC^
C^
Suffix Protein Fusion Preparation Part (BioBrickPF-S): Composition of a BioBrick protein part and a BioScaffold part that can be cut with the BioScaffold part specific enzyme, leaving characteristic ends.
Table 8: Format of the BioBricPF-S (BioScaffold and protein composite) parts BioBrickPF-S(Type1) Part:(Suffix Protein has a head, BioScaffold specific enzyme is PsrI) <Prefix w/AG> BBa_J70012 <Scar w/AT>G GGATCC protein TAA<Suffix> BioBrickPF-S(Type1) Part:(Suffix Protein lacks a head, BioScaffold specific enzyme is PpiI) <Prefix w/AG> BBa_J70010 or BBa_J70030 <Scar w/AG> GATCC protein TAA<Suffix>
Table 9: Format of the BioBrickPF-S parts after excision with the BioScaffold specific enzyme:
^ protein TAA<Suffix>
^CTAGG
The current BioScaffold parts enable automated 3A assembly using BioBrickPF-P and BioBrickPF-S parts since they can be heat killed and removed by Qiagen PCR spin columns or other purification techniques depending on the size of the proteins []. This allows both the BioScaffold parts and appropriate BioBrick ends on the prefix and suffix proteins to removed so that the the protein fusion parts can be ligated into a vector containing a different antibiotic resistance than the original BioBrickPF-P and BioBrickPF-S part containing vectors.
Table 10: Composing BioBrickPF-P and BioBrickPF-S parts (using 3A assembly and after excision of the
BioScaffold parts)
Assembly:
<EcoRI cut Prefix> protein G GATCC^ + ^ protein TAA <PstI cut Suffix>+<EcoRI and PstI cut Vector>
C^ ^CTAGG
Final construct:
<Prefix> protein GGA TCC protein TAA <Suffix> <Vector>
CCT AGG
Gly-Ser
The Library Construction Problem
The goal is to be able to introduce new parts (often with a variable region) into an assembly as the last step in the construction process.
Library Construction Using BioScaffold β parts
Cut within the scars
BioScaffold Interior Parts
^pppp pppp GCAGGTG GACAAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC ssss^ssss
pppp^pppp CGTCCAC CTGTTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG ssss ssss^
AarI site XmaI site AarI site
Vector Preparation
<Prefix> part1 <Scar> BBa_JX <Scar> part2 <Suffix>
<Prefix> part1 TACTAGAT BBa_JX TACTAGA(T or G) part2 <Suffix>
ATGATCTA ATGATCT(A or C)
<Prefix> part1 ^TACT AGAT BBa_JX TAC^TAGA (T or G) part2 <Suffix>
ATCA^TCTA ATG ATCT^(A or C)
<Prefix> part1 ^ ^TAGA (T or G) part2 <Suffix>
ATCA^ ^(A or C)
BBa_JX= BBa_J70034
^TACT AGAT GCAGGTG GACAAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC A TAC^TAGA
ATGA^TCTA CGTCCAC CTGTTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG T ATG ATCT^
AarI site XmaI site AarI site
<Prefix w/AT or AG> GCAGGTG GACAAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC A <Suffix>
CGTCCAC CTGTTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG T
Insert Preparation <Prefix> BBa_JY <Scar> insertpart <Scar> BBa_JY <Suffix>
<Prefix> BBa_JY TACTAGA(T or G) insertpart TACTAGAG BBa_JY <Suffix>
ATGATCA(A or C) ATGATCTC
<Prefix> BBa_JY ^TACT AGA(T or G) insertpart TAC^TAGA G BBa_JY <Suffix>
ATGA^TCA(A or C) ATG ATCT^C
^TACT AGA(T or G) insertpart TAC^
^TCA(A or C) ATG ATCT^
The vector and the insert or inserts can now be ligated together to create the final part.
<Prefix> part1 ^TACT AGA(T or G) insertpart TAC^TAGA (T or G) part2 <Suffix>
ATCA^TCA(A or C) ATG ATCT^(A or C)
BBa_JY
TAC^TAGA (G or T) ATA GCAGGTG GACAAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC ATAT ^TACT AGA(G or T)
ATG ATCT^(C or A) TAT CGTCCAC CTGTTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG TATA ATGA^TCT(C or A)
<Prefix> ATA GCAGGTG GACAAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC ATAT <Suffix>
TAT CGTCCAC CTGTTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG TATA
tactagat ^TACT AGAT PI1 TAC^TAGA (T or G)
ATGA^TCTA ATG ATCT^
tactagag ^TACT AGAG PI1 TAC^TAGA (T or G)
ATGA^TCTC ATG ATCT^
will these anneal? no
^ ^TAGA
ATGA^ ^
Preparation parts
(PI2 Left) ^TACT AGAT library insert TAC^TAGA (G) (PI3 Right)
ATGA^TCTA ATG ATCT^(C)
Another way but may anneal
TA^CTAG AT library insert ^TACT AGA(G)
AT GATC^TA ATGA^TCT(C)
For parts preparation
TACTA^GAG B1 TA CTAGA^G ^ATGAT CTC part to remove AT^GATCA C
TACTA^ ^ ^ ^GATCA
For final introduction
TA CTAGA^G Part ^TACTA GAG AT^GATCA C ATGAT^CTC
<Prefix><Psr I><Construct><Scar w/AG><PpiI><Scar w/AG><PsrI><Suffix> <Prefix><Psr I><
B2left TACTA^GAG part to insert TA CTAGA^G
^ATGAT CTC AT^GATCT C
Library Construction Using BioScaffold α parts
First place a BioScaffold part between two BioBrick parts as shown in Table 2.
You do not continue directly along the lines of the protein fusion protocol for parts designers, because the digestion efficiency of the BioScaffold specific enzymes PpiI and PsrI are below 95 percent which is not ideal for library construction, where you wish to retain as much diversity (or complexity in the library as possible) and this inefficiency would reduce the number of clones produced.
Remove the BioScaffold part as appropriate for that part.
<Prefix> . . .ppp ppppp^ ^ssss. . . <Suffix>
ppp^ ^Cssss ssss
Replace it with a Peisajovich inspired BioMortar part (this type of part is inspired by Sergio Peisajovich's AarI assembly standard developed with the UCSF 2007 iGEM team and Wendell Lim.) Then sequence to select appropriate clones.
Peisajovich inspired BioMortar part (Melting Temperature greater than 76 degrees Celsius)
5'- GCAGGTC GACAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC Gssss-3'
3'-ppppp CGTCCAC CTGTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG -5'
AarI site XmaI site AarI site
<Prefix w/AT>G . . . ppppp GCAGGTC GACAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC Gssss . . . TAA<Suffix>
ppppp CGTCCAC CTGTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG Cssss
Digest with AarI and XmaI and purify according to the Peisajovich AarI digestion protocol [], yielding the following product.
<Prefix w/AT>G . . .^pppp pppp GCAGGTC GACAGAGGAGT CCCGGG AGCTGGAACTCC CACCTGC Gsss^ssss s . . . TAA<Suffix>
pppp^pppp CGTCCAC CTGTCTCCTCA GGGCCC TCGACCTTGAGG GTGGACG Csss ssss^s
The Parts Domestication Problem
Currently, it is not yet clear whether nonstandard BioBrick parts will be sequenced, maintained, or distributed by the Registry of Standard Biological parts in the long term. Nonstandard parts are problematic for the registry because they impose additional costs on the user. However, in cases when the BioBrick standard could not meet all the needs of the user, nonstandard parts have sometimes been allowed. BioScaffold parts, by building upon the work of the community, suggest a way in which the need for these nonstandard parts can be minimized.
nonstandard parts are not allowed in the registry. This is good because it means that parts can be used interchangeably and that the parts included in the Registry distribution are high quality parts.
However, sometimes there are reasons why a BioBrick parts user, especially a parts designer or someone from outside the synthetic biology community, may not want to immediately convert a part to the BioBricks standard.
-There is not yet a one step protocol for converting more than 5 restriction in a part over to the BioBrick standard though (http://openwetware.org/wiki/Knight:Site-directed_mutagenesis/Multi_site provides a protocol for up to 5 sites at a time.) Creating a process that routinely achieves conversion in one step for up to 10 sites would be an excellent BBF RFC standard.
-They may be publishing a paper and want to direct people who want the part to the Registry both to share their part and in order to attract more citations for their paper, however, they do not want to delay publication of their paper.
-For some technical/functional reason converting their part to the BioBrick standard does not make sense. In this case the reason should be reported to the community in a BBF RFC so that the community can try to find a way to fix the problem.
-They wish to donate a large collection of parts to the Registry.
-They would like to try out a new standard for a certain part of the assembly process and have not yet found a way to integrate their new standard with the BioBricks standard (we hope that the new BioScaffold parts will help facilitate integration.)
Philosophy
Reasons to allow non-BioBrick parts into the registry
-Registry and collection of parts will grow more quickly
-With BioScaffold parts, non-BioBricked parts can be placed in BioBricked vectors with minimal effort such that they can be sequenced, maintained, and distributed by the Registry
-BioScaffold parts now make it possible to integrate non-BioBricked parts into BioBricked constructs (as described below) -Allows parts designers greater freedom in manipulating parts and testing options before selecting the optimal part
-Divergent standards will not cause fragmentation of the community due to different parts only being held in multiple divergent registries
-Paper authors can submit their non-BioBricked parts to the registry before publication to satisfy materials distribution requirements and attract additional citations to their papers, however at the same time their path to publication will not be slowed
However non-BioBrick parts impose unnecessary costs on the user, and across the whole community the costs become significant
-incompatible with existing parts since do not allow standard assembly
-require additional sequencing (each part must be sequenced fully when prepared by pcr, rather than just be verified)
-lead to storage of more instances of the part in the registry
-the BioScaffold enzymes are not as efficient as the BioBrick enzymes, and manipulations with them are more time consuming
Thus, the community should incentivize conversion from BioBrick parts to non-BioBrick parts as soon as possible A few possibilities -Create a non-standard BioBrick part sandbox, maintained by the Registry-- parts in this sandbox will not be distributed until someone claims the conversion rights for the part-- they then have 6 months to convert the part and return it to the registry
-Create a BioBrick part journal as part of the parts promotion process, and not allow publication until conversion -After conversion, credit both the part creator and part converter as authors in the BioBrick parts journal -Allow people to claim conversion rights on the registry's
B-S enzymes available
PsrI, PpiI, AarI
<BS
Always want to add your undomesticated parts last to the assembly?
<BS-Non-Cutter> <Part> <Part> <Part> <Part><BS Cutter><BS-Non-Cutter>
<BS-Non-Cutter> <Part> <Part><BS-Cutter><Part> <Part><BS-Non-Cutter>
<BS-Non-Cutter><BS-Cutter> <Part> <Part><Part> <Part><BS-Non-Cutter>
<Prefix Scar>
<BS-Non-Cutter><Scar><UD Part><Scar><BS Non-Cutter>
<Scar Suffix>
<Scar Scar>
<Scar> tactagag or tactagat(g)
<Prefix> gaattcgcggccgcttctagag or gaattcgcggccgcttctagat(g)
<Suffix> tactagtagcggccgctgcag
Allowing a construct to be transferred to a new vector, while removing the BioScaffold parts (<B-P> parts)
Old vector
<Prefix> <B-P><Scar> construct<Scar> <B-P> <Suffix>
gaattcgcggccgctt ctaga g ta ctaga^(t or g) tacta^gag tacta gtagcggccgctgcag
^ ^
New vector
<Prefix> <B-P part sealer>
gaattcgcggccgctt ctaga^g tacta^gtagcggccgctgcag
^ ^
<Prefix> <B-P1 A> <Scar> construct <Scar> <B-P2 A> <Suffix>
gaattcgcggccgc ttcta^gag ta ctaga^(t or g) tacta^gag ta ctagt^agcggccgctgcag
^ ^ ^ ^
Left part vector insertion vector <Prefix><B-P3 A><Scar>xx ttcta^gag<B-P1 A><Scar> <B-P beta> <Scar><B-P3><Suffix>
Right part vector insertion vector <Prefix><B-P4><Scar><B-P beta><Scar><BP-2>ta ctagt xxx <Scar><BP-4><Suffix>
Part insertion pcr format <B-P beta><Scar>part<Scar><B-P beta> Part insertion oligo format XXXXXXXXX
<BioScaffold PsrI> <Scar> construct <Scar> <BioScaffold PpiI> <Scar> <BioScaffold PsrI>
<BioScaffold PsrI> <Scar> Part <Scar> <BioScaffold PsrI>
<BioScaffold PsrI> <Prefix> construct <Scar> <BioScaffold PpiI> <Suffix> <BioScaffold PsrI>
<BioScaffold PsrI> <Scar> Part <Suffix> <BioScaffold PsrI>
Selected BioScaffold Parts and Examples of their Use
To demonstrate the utility of BioScaffold parts the current designed BioScaffold parts in the Registry (Figure 7) and examples of their potential use are included (Figure 8).
| Figure 7: Examples of BioScaffold Parts | ||
|---|---|---|
 |
 |
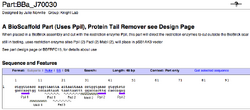 |
| Figure 8: How They Work | ||
 |
 |
 |
References:
[1] Julie E. Norville and the Synthetic Biology Community, "BBFRFC15 Comments Page," The BioBricks Foundation, December 30, 2008, <http://www.openwetware.org/wiki/The_BioBricks_Foundation:BBFRFC15_Comments_Page>.
[2] Julie E. Norville and the Synthetic Biology Community, "BioScaffold Parts," OpenWetWare, December 30, 2008, <http://openwetware.org/wiki/BioScaffold_Parts>.
[3] Julie E. Norville and the Synthetic Biology Community, "BioScaffold Parts Users' Group Wiki," OpenWetWare, December 30, 2008 <http://www.openwetware.org/wiki/BioScaffold_Parts_Users_Group>.
[4] Tom Knight, Randall Rettberg, Leon Chan, Drew Endy, Reshma Shetty, and Austin Che "Idempotent vector design for standard assembly of BioBricks," BBF RFC 9, 2003, <http://openwetware.org/wiki/Media:BBFRFC9.pdf>.
[5] Tom Knight, "Draft standard for BioBrick biological parts," BBF RFC 10, May 3, 2007, <http://openwetware.org/index.php?title=The_BioBricks_Foundation:BBFRFC10&oldid=262187>.
[5] Tom Knight, "Draft BB-2 standard for biological parts," BBF RFC 12, November 19, 2008, <http://openwetware.org/index.php?title=The_BioBricks_Foundation:BBFRFC12&oldid=263570>.
[6] Tom Knight, "Rethinking the boundaries and composition of coding regions," BBF RFC 13, November 19, 2008, <http://openwetware.org/wiki/BBRFC13>.
[7] Tom Knight, "Protein domain fusions in BB-2 assembly," BBF RFC 14, November 23, 2008, <http://openwetware.org/wiki/BBRFC14>.
BBFRFC9 [9], BBFRFC12 [10], BBF RFC13 [11], BBF RFC14 [12],
[1] Tom Knight, Randall Rettberg, Leon Chan, Drew Endy, Reshma Shetty, and Austin Che "Idempotent vector design for standard assembly of BioBricks," BBF RFC 9, 2003, <http://bbf.openwetware.org/RFC.html.>
[2] Tom Knight, "Draft standard for BioBrick biological parts," BBF RFC 10, May 3, 2007, <http://bbf.openwetware.org/RFC.html.>
[3] Austin Che, "BioBricks++: Simplifying Assembly of Standard DNA Components," June 9, 2004, <http://austinche.name/docs/bbpp.pdf>
[4] Ira Phillips and Pamela Silver, "A New Biobrick Assembly Strategy Designed for Facile Protein Engineering," MIT SBWG Technical Reports, April 20, 2006, <http://dspace.mit.edu/handle/1721.1/32535>
[5] Tom Knight, "Biobrick assembly standard modification," BBF RFC 11, July 8, 2008, <http://bbf.openwetware.org/RFC.html.>
[6] Tom Knight, "Draft BB-2 standard for biological parts," BBF RFC 12, November 19, 2008, <http://bbf.openwetware.org/RFC.html.>
[7] Tom Knight, "Rethinking the boundaries and composition of coding regions," BBF RFC 13, November 19, 2008, <http://bbf.openwetware.org/RFC.html.>
[8] Tom Knight, "Protein domain fusions in BB-2 assembly," BBF RFC 14, November 23, 2008, <http://bbf.openwetware.org/RFC.html.>
[9] BioBricks Foundation: Standards/Technical/Formats <http://openwetware.org/wiki/The_BioBricks_Foundation:Standards/Technical/Formats#The_Berkeley_.28BBb.29_Format>
[10] Sergio Peisajovich, Wendell Lim, and 2007 UCSF iGEM team, October 30, 2008, <http://2008.igem.org/Team:UCSF/Materials_and_Methods.>
[11] Chris Anderson and Zhan Jian, Plasmids/Library Screening, <http://partsregistry.org/Plasmids/Library_screening.>
[12] Reshma Shetty and Randall Rettburg, "3A Assembly," December 13, 2007, <http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly.>
[13] Kristian Müller and Katja Arndt, "Restriction sites for the construction of fusion proteins," BBF RFC 3, <http://bbf.openwetware.org/RFC.html.> (Note: this RFC is not yet posted, but is expected to describe the standard in [9].)
[14] Julie Norville, "Part: BBa_J70010, A BioScaffold Part," Registry of Standard Biological Parts, May 5, 2008 <http://partsregistry.org/Part:BBa_J70010.>
[15] Julie Norville, "Part: BBa_J70012, A BioScaffold Part," Registry of Standard Biological Parts, May 5, 2008 <http://partsregistry.org/Part:BBa_J70012.>
[16] Julie Norville, "Part: BBa_J70030, A BioScaffold Part," Registry of Standard Biological Parts, December 12, 2008 <http://partsregistry.org/Part:BBa_J70030.>
[17] Randall Rettberg and others, "Help: BioBrick Prefix and Suffix," Registry of Standard Biological Parts, July 23, 2006 <http://partsregistry.org/Help:BioBrick_Prefix_and_Suffix.>
[18] Reshma Shetty, "Plasmid Backbones/Archive," Registry of Standard Biological Parts, December 11, 2008 <http://partsregistry.org/Plasmid_backbones/Archive.>
[19] Reshma Shetty, "Plamid Backbones/Assembly," Registry of Standard Biological Parts, December 11, 2008, <http://partsregistry.org/Plasmid_backbones/Assembly.>