The Actin Cytoskeleton of the Cell by Laura Feeley and Jeremy Keys
Background
Cytoskeleton Overview
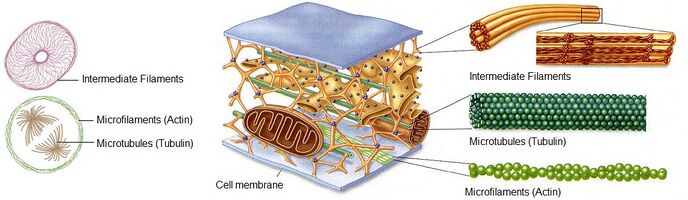
The cytoskeleton is an array of proteins in the cytoplasm that form the structural framework of all eukaryotic cells and most prokaryotic cells. The cytoskeleton is composed of three primary components: microtubules, actin microfilaments, and intermediate filaments. Each of these components provides a specific set of functions to regulate the overall behavior of the cell. Microtubules (dark green), composed of tubulin, are straw shaped structures which are approximately 20 nm in diameter and are the largest of the cytoskeletal components. Microtubules are involved in mitosis, cell movement, intracellular transport and cell structure. Actin filaments (light green) are the smallest of the three components and are composed of a helical strand of actin monomers averaging around 6 nm in diameter. Actin will be further discussed. Intermediate filaments (orange) are twisted groups of proteins which provide tensile strength to the cell and are 10-20 nm in diameter. Intermediate filaments are less dynamic than actin and microtubules and primarily provide support for the cell. Actin and microtubules are similar in that they are polar structures with constant additions and subtractions to specific ends. Collectively, the cytoskeleton works to control cell shape, add structural strength, produce motility, and aid in cell division.
Actin
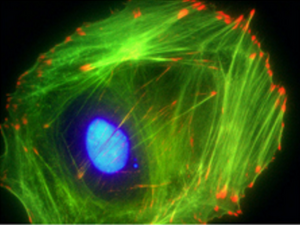
Actin is the most plentiful intercellular protein in eukaryotic cells, accounting for 10% of cellular protein in muscle cells and 1-5% in most non-muscle cells. Initially recognized for its role in muscle contraction, actin is a primary component of the cellular cytoskeleton and plays a key role in cell motility, cell division, and cell-surface and cell-cell interactions.
Actin exists primarily in two forms: filamentous (F-actin) and globular actin (G-actin). Under physiological conditions, as much as 99% G-actin will naturally polymerize into F-actin [2]. Actin is often classified among three isotypes: alpha-actin, beta-actin, and gamma-actin. Alpha-actin is found primarily in muscle cells, being responsible for muscle contraction along in cooperation with myosin, while beta-actin and gamma actin are regularly associated with the cytoplasm of non-muscle cells. In addition to these three isotypes, there are numerous small variations on actin structures between organisms. While there is relatively little diversity among actin's isotypes, actin can observe a broad array of behaviors due to its interactions with actin-binding proteins (ABPs). As over 150 ABPs have been identified (which accounts for 25% of cellular proteins), actin's functions within the cytoskeleton are very diverse.
A linear actin polymer is the basis of all cytoplasmic actin. Two actin strands are non-covalently bound to each other in a helical shape. The actin polymers are made up of actin monomers (Figure 2). These polymers are synonymous with F-actin whereas G-actin indicates only the monomeric actin form. A strand with greater than 6-7 monomers allows the actin polymer to bend[11]. The helix is formed with 13 monomers, six left handed turns and an overall diameter of 5-9nm [11]. The actin protein itself has a globular shape with 4 subunits. The joining of the subunits in the middle yields an ATP or ADP binding site. Binding of different states yields changes in the structure of the protein. Focusing on the filamentous actin, it is a polar structure which has a negative (or sharp) end and a positive (or barbed) end.
Actin Polymerization
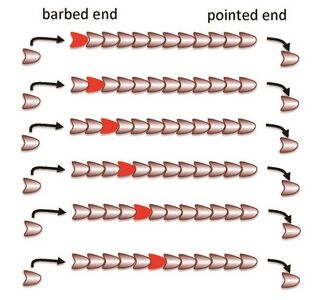
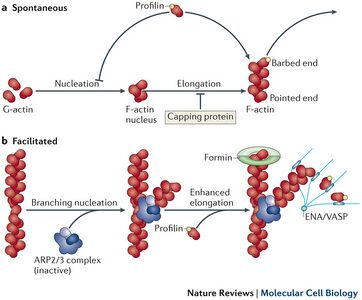
Three phases of actin polymerization have been identified: lag period, elongation phase and equilibrium phase [11]. Actin polymerization begins with a collection of actin monomers (G-actin) called a “nucleus.” The polymerization process begins with stabilization of the nucleus by “nucleation,” before the elongation of the polymer. The addition of monomers yields a growth rate of about 1 micrometer/second [11]. In polymerization, monomers are being added to both the sharp and barbed end but addition is primarily favored on the barbed (positive) end and disassociation is primarily found at the pointed (negative) end. After the elongation, the filament reaches equilibrium with monomer concentration. This concentration of monomer, critical concentration, determines if addition to or dissociation of the filament will occur. The addition and disassociation of actin from subsequent ends of the polymer yields a phenomenon called treadmilling [11]. Treadmilling can be seen when a monomer is added to the barbed (positive) end and ultimately moves down the polymer until it eventually disassociates at the pointed (negative) end (Figure 3). This polarity of the polymer is what drives forward motion of the cell. The role of actin binding proteins on polymerization also has a significant impact on how quickly a cell is able to move. As previously mentioned, polymerization requires nucleation, which needs nucleators. A common nucleator is, actin related protein complex 2/3 (ARP2/3) [11]. This actin binding protein can also bind to existing filaments and cause branching of the actin filament at each of its binding sites [4]. Each branch consists of a Y-shape at a 70 degree angle from the original filament (Figure 4)[11]. By branching the actin, a second barbed end is formed and therefore the polymerization rate may be doubled provided there is enough G-actin available. Cofilin is another actin binding protein which produces a similar effect by splitting the polymer in half, thus producing a second barbed end and increasing the polymerization rate [4]. For a closer look at actin polymerization and the effects of binding proteins Arp2/3 and Cofilin, an Actin Polymerization video from the Mechanobiology Institute at the National University of Singapore can be viewed. Actin polymerization can also be affected by capping proteins that bind either to the barbed or pointed end. An example of a capping protein is CapZ, which binds to the barbed end and prevents polymerization [11]. This in turns shortens the polymer because disassociation is occurring on the pointed end.
History of the Actin Cytoskeleton
While actin’s presence in muscle cells was understood as early as the late 19th century and the idea of a structural framework for cells was proposed as early as 1903, the role of the actin in the cytoskeleton of eukaryotic cells was not well understood until the advent of fluorescent labeling techniques in the early 1970s. A brief summary of key discoveries regarding the actin cytoskeleton include the following:
- 1887- Actin is first purified by W.D. Halliburton
- 1950- Role of Actin in muscle contraction first proposed (Straub)
- 1953- Earliest studies of cell motility (Abercrombie)
- 1968- Actin filaments observed in non-muscle cells (Holtzer, Ishikawa)
- 1970- Actin first implicated in cell motility (Wessels, Spooner, Yamada)
- 1974- Actin network is first visualized in non-muscle cells (Lazarides, Weber)
- 1980- Reproduction of actin cytoskeleton through “freeze-etching” (Heuser)
- 1992- Role of Rho GTPases in cytoskeletal regulation (Hall)
- 1995- First actin mutation is associated with skeletal muscle myopathy (Liang)
- 1999- Reconstruction of in vivo complete motile system (Louisel, Carlier). [5]
Cell Motility
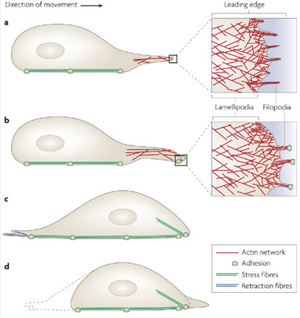
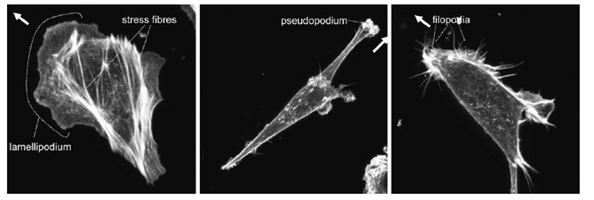
The most significant role of the actin cytoskeleton is its ability to control cell locomotion. Cell motility is driven by the rapid polymerization of actin filaments at the front of the cell coordinated with the gradual disassembly of actin filaments towards the cell rear. The polarization of the filament is what gives motility directionality. Polymerization at the front of the cell, which is driven by Rho GTPases, causes a large forward protrusion of a “lammelipodium” [2]. The lammelipodium is formed as forward extension of actin proteins stretches out the cell membrane. The lamella is posterior to the lammelipodium part of the protrusion [11]. The actin filaments in the lamella are less branchlike but thicker than the lammelipodium. The actin fibers are coated in Myosin II which aids in the contractile motion of the cell. Smaller, finger-like protrusions called “filopodia” then form focal adhesions with the substrate beneath the lammelipodium [6]. Overall cell motility occurs when the lammelipodium is formed due to polymerization at the leading edge of the cell(a). The filopodia then form focal adhesions with the substrate surface (b). The increase in cell tensions activates LIM-proteins to aid in reorganizing actin fibers into bundles to function as stress fibers(c). The cell tension then breaks focal adhesions at the cell rear and thus pull the cell forward(d).[6]
The role of Cell Motility in Cancer Metastasis
Metastasis, or the spread of cancer from one location of the body to another, is dependent on the ability of its cells to migrate throughout the body. In order to accomplish this, cancer cells are highly reliant on the cytoskeletal processes of motility in order to intravasate (or invade) other tissues. It has been demonstrated in some strains of breast cancer cells that the production of ARP2/3 and cofilin are upregulated while Eva/VASP proteins are shut down [4]. Eva/VASP proteins are associated with the formation of filopodia. Therefore, these cancer cells effectively increase the rate of polymerization to push forward lamellipodia faster, and eliminate the formation of filopodia in order to increase the surface area which the front of the cell attaches to; each of these results in a much faster moving cell. While not all cancers modify the regulation of the cell's motile mechanisms in the same way, many change these structures in some way in order to improve their chances of survival and spreading.
In addition, cancer cells hijack the ability to form actin-driven protrusions in order to form specialized protrusions called "invadopodia" [4]. Invadopodia are crucial for metastasis because they push into the surrounding extracellular matrix (ECM) to open up holes, and also work to break down the ECM [4]. For cancer cells which are contained in otherwise isolated regions of the body, the formation of invadopodia are their means of spreading into other tissues.
Potential Actin Cytoskeleton Targeted Therapies for Cancer
Cancer
In 2013, a group of researchers in New South Wales, Australia led by Dr. Peter Gunning and Dr. Justine Stehn completed several successful animal trials of a drug which targets tropomyosin, an actin binding protein associated with motility, in tumor cells [9]. By affecting the cells motility, the ability of tumor cells to seek out nutrients and spread throughout the body is significantly reduced, ideally leading to the cells starving themselves. The drug, named TR100 was slated to enter human trials in a group of 12 children suffering from advanced neuroblastoma in 2015. An advantage of the drug is that it is potentially applicable to many types of cancer because it targets structures which is necessary for all cancer cells to thrive.
Challenges
Drugs which target the cytoskeleton in cancer cells have been investigated in the past, but the difficulty with such drugs is controlling them in such a way that they exclusively impact cancer cells. Tropomyosin is critical for the function of many cells in the body, particularly in skeletal and cardiac muscle. It is for this reason that most researchers have not continued investigation of targeting cancers in this manner [9]. The researchers have stated that they will be closely monitoring the muscular and cardiac health of their patients, however it was notable that these negative side effects were not observed in their animal models. [9]
Drugs Targetting Actin Polymerization
Three classes of pharmalogical drugs
Three main classes of drugs have been studied to inhibit actin polymerization.
The first class of drugs interfere with actin polymerization. Cytochalasin D inhibits elongation at the barbed (positive) ends of the filament while Latrunculin inhibits monomers aggregation and therefore stops polymerization [12].
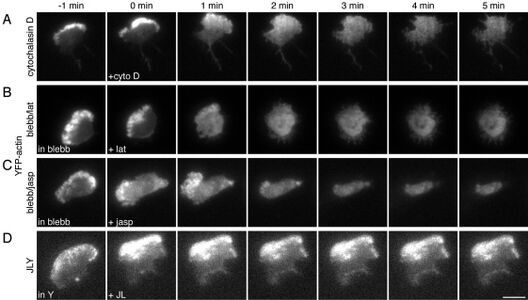
The second class of drugs interfere with the disassembly of the polymer. Jasplakinolide is a cyclic peptide derived from a marine sponge that aims to stabilize the actin filament, inhibit depolymerization and promote polymerization [13]. It also minimizes the dissociation rate constant for the barbed end which ultimately causes the incorporation of all free actin [12]. Side effects of this drug caused cells to develop malformed patterns, changes in cell shapes, abnormal protrusions in cells and binucleated cells when it was applied during mitosis [12]. In vitro tests displayed that Jasplakinolide was successful in stabilizing actin filaments but in vivo testing induced polymerization of monomeric actin into amorphous masses [13].
The third class of drugs aimed to interfere with the rearrangement of the network in order to favor contraction rather than protrusion. Myosin II is important in disassembly of the entire actin network. The third class of drugs includes blebbistain which inhibits Myosin II and Rho kinase (ROCK) inhibitor which inhibits an activator for Myosin II. A common drawback to each of these drugs were related to toxicity levels and the death of surrounding cells.
Triple-drug Cocktail to inhibit actin dynamics
A triple-drug cocktail containing jasplakinolide, latrunculin B, and Y27632, (JLY) was created to inhibit actin depolymerization, actin polymerization, and myosin II-based contractility respectively, without influencing polymer assembly, disassembly, or rearrangement [14]. One issue with these drugs individually is that although they can be effective, subsequent rearrangement occurs. In this study HL-60 cells were chosen because they displayed neutrophil-like behaviors in regards to rapid actin polymerization [14]. Specific doses of the JLY cocktail were found to “freeze” cell morphology which means that no polymerization, depolymerization or rearrangement occurred. JLY was also found to keep the polymer at steady state through no additions or deletions of monomers. The goal of this cocktail was to incorporate the effective aspects of the three classes of drugs but apply them in a way that did disturb surrounding cells through polymerization, depolymerization or rearrangement. When compared with blebbistain, and cytochalasin D, the researchers found that the JLY cocktail kept the actin cell morphology intact and preserved the steady state actin (Figure 7)[14].
Disruption of the actin filament through targeting isoforms of tropomyosin
Recently research has shifted to developing drugs that target regulatory proteins rather than targeting actin directly, due to previously mentioned toxicity issues. These actin targeting drugs cannot specifically target actin in tumor cells and therefore are very toxic to healthy cells. In 2013, researchers from the University of New South Whales, began targeting isoforms of tropomyosin as a method for actin filament disruption [15]. Tropomyosin is an important coiled -coil protein within actin filaments that is located within or along the alpha helix where it interacts and regulates the filament[15]. As mentioned, in the cytoskeleton there are two possible actin filament isoforms, and over 40 tropomyosin isoforms [15]. The researchers developed anti-tropomyosin compounds and chose TR-100, since it was deemed most effective. This work was unique in that it deterred tumor growth but did not effect cardiac function in vivo. Targetting tropomyosin can often harm cardio and skeletal muscle. They identified a particular isoform, Tm5NM1, that was continuously found in the tumor cells they were analyzing. Since Tm5NM1 reduced F-actin depolymerization, they studied the rate of depolymerization in F-actin and in F-actin coated with Tm5NM1 [15]. TR100 eliminated the reduction of polymerization caused by Tm5NM1, therefore displaying it's efficient inhibitory abilities. Studies were conducted in both neuroblastoma and melanoma cancer cells. TR100 had a significant impact on actin but did not disrupt the microtubules within the cytoskeleton. There is not clear evidence that TR100 can specifically target low or high molecular weight tropomyosins, so this will be further researched. It was also determined that TR100 would preferentially target actin cytoskeleton filaments rather than muscle actin filaments [15].
Diseases caused by Actin binding protein Defects
The actin cytoskeleton plays important roles in all aspects of a functioning immune system; hempatopoiesis, immune cell development, recruitment, migration, intercellular signaling, and activation of immune responses [16]. Due to this role, there are many actin linked primary immunodeficiencies (PIDs)
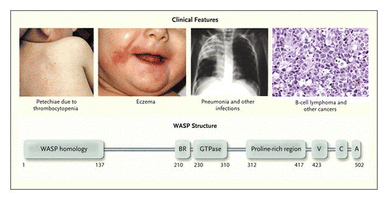
Wiskott-Aldrich syndrome (WAS)
WAS is an X-linked recessive disorder first identified in 1937. Symptoms of WAS are low platelet count, eczema, bloody diarrhea, recurrent fever, and ear infections (Figure 8)[16]. The gene that is responsible for this disorder is on chromosome 25 which controls the WASp family that regulate ARP 2/3, a common actin nucleator. There are over 300 possible disease causing mutations in the WAS gene that cause specific disorders. Classic WAS is caused by complete loss of WASp function. X-linked thrombocytopenia (XLT), deficiency of platelets in blood causing bleeding in tissues and slow clotting, is caused by mutations that reduce WASp expression. X linked neutropenia, or low levels of neutrophils, is caused by a mutation within the GTPase binding domain (GBD) disrupting the auto-inhibited conformation of WASp and yielding an active protein [16]. Mutations in the WASp family can cause a variety of disorders.
DOCK8 deficiency
Dedicator of Cytokinesis 8 (DOCK8) are found in the edges during lamellipodia formation in cell motility and aid in regulation of actin polymerization kinetics [16]. DOCK 8 deficiencies have been found to cause by severe food and environmental allergies, otitis media, pneumonia, bronchitis, eczema, and severe cutaneous viral infections such as human papilloma virus (HPV), herpes and varicella-zoster virus. The DOCK8 deficiency is linked to an autosomal recessive form of hyper IgE syndrome (AR-HIES). AR-HIES is an immune system disorder with increased susceptibility to infections that can lead to mortality. Zhang identified point mutations in DOCK8 in patients suffering with AR-HIES [16].
CORO1A mutation
Coronins are a family of actin regulatory proteins. Type I Coronins regulate actin polymerization in the leading edge of the cell through inhibiting nucleation of monomers in cells with a large degree of branching [16]. Nucleation is directly inhibited through the inhibition of Arp2/3 [16]. Coronin1A is commonly found in T cells. A mutation in type I coronins causes severe immunodeficiency, T-cell deficiency and susceptibility to EBV-induced lymphoproliferation, where lymphocytes are excessively produced. In studying mice cells with lupus, the suppression of type I coronin, Coronin 1A, seemed to inhibit all immune responses [16].
Actin-accumulation myopathy
Actin found in muscle cells is also linked to specific disorders.
Actin-accumulation myopathy is found in skeletal muscles where the patient experiences severe muscle weakness and tone. This is usually first seen in infants who have trouble feeding, swallowing, controlling head movements, and breathing. Most patients with disease d not survive past infancy and if they do, delayed motor skills are usually observed.
Other Applications for Actin targeted therapies
Gene Editing for Dystrophy

Genetic defects to actin which cause dystrophies have had no comprehensive treatments in the past, however recent mouse trials utilizing adeno-associated virus (AAV) delivery were successful in recovering the missing gene necessary for the formation of dystrophin (actin binding protein which is missing in patients suffering from dystrophy) [10]. If these processes can be shown to continue to be successful, gene editing could represent a major change in how dystrophies are treated.
Ethical Concerns
Gene editing has faced considerable controversy due to its potential applications outside of treating disease. Specifically, many are concerned with the ethical ramifications of a tool which might be used towards "improving" humans; treating conditions or modifying ones genes in order to reach a biological ideal. These, of course, have associations with ideas such as eugenics which has put the idea of gene editing under scrutiny.
Other factors impacting gene editing's success include a group of 3 deaths which occurred during human trials of gene editing treatments between 1999 and 2007. While there is some debate over whether or not all of these deaths were actually a result of the treatment, skepticism towards its viability as a treatment has risen.
Additionally, due to the lack of human testing of gene editing, there is still little known about the long term effects of gene editing in humans. Other diseases associated with specific mutations can be found in Figure 9.
Actin Polymerization Kinetics
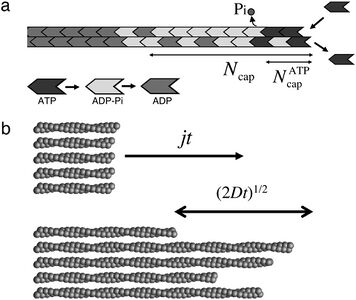
The study of actin polymerization has been a popular subject for decades. Recent research has focused on developing kinetics models of actin polymerization with the application of a specific drug. Researchers at Columbia University are focusing on analyzing a single filament with non-steady state growth rates accounting for monomers added to the growing filament as ATP-actin, ATP being hydrolyzed to ADP and phosphate (Pi) and the release of Pi into the solution (Figure 10)[17]. Kinetic research frequently focuses on long actin chains consisting of ADP-actin so the dynamics of this model is unique. They concluded through their model that growth rates and the critical concentrations on the barbed end are closely related to cap structure and kinetics of actin polymerization [17]. The cap structure refers to a three state region on the end of the filament that can regulate additions to the filament. Changes in filament lengths were measured by length diffusion coefficient D, in terms of ATP-actin concentration. The polymer length fluctuated at concentrations above the critical concentration, which is the amount of monomers present when the filament is in steady state. The researchers attributed this to growth being controlled by oligomer dynamics rather than critical monomer concentrations [17]. Further modeling of actin polymerization kinetics could provide a novel approach controlling cell motility in cancer cells.
References
[1] "Actin Staining Techniques." Cytoskeleton Inc. Cytoskeleton Inc, 2015. Web. 29 Feb. 2016.
[2] Hall A., (1998), Rho GTPases and the Actin Cytoskeleton. Science. 279: 509-514.
[3] Holmes K.C. (2009), Structural Biology: Actin in a Twist. Nature. 457: 389-390
[4] Olson MF, Sahai E (2009), The actin cytoskeleton in cancer motility, Clin Exp Metastasis, 26:273-287
[5] "Cytoskeleton Milestones Timeline." Nature Milestones. Macmillan Publishers Limited, 2016. Web. 29 Feb. 2016.
[6] Smith M.A., Hoffman L.M., and Beckerle M.C. (2014), LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol., 24: 575-583.
[7] Rubenstein P.A., Wen K.K., (2014), Insights Into the Effects of Disease-Causing Mutations in Human Actins. Cytoskeleton, 71:211-229. Doi: 10.1002/cm.21169
[8] Rahminov F and Kunkel L.M. (2013), Cellular and molecular mechanisms underlying muscular dystrophy. J. Cell Biol. 201: 499-510.
[9] Stehn JR, Haas NK, and Bonello T et al. (2013), A Novel Class of Anticancer Compounds Targets the Actin Cytoskeleton in Tumor Cells. Cancer Res. 73: 5169
[10] Tabebordbar M, Zhu K, and Cheng JK et al (2015), In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. doi: 10.1126/science.aad5177
[11] Lee, J. (2013). The actin cytoskeleton and the regulation of cell migration. Colloqium series on building blocks of the cell: cell structure and function. [
[12]Bubb, M., Spector, I., Beyer, B., & Fosen, K. (2000). Effects of Jasplakinolide on the kinetics of actin polymerization. The Journal of Biological Chemistry, 275, 5163-5170.
[13] Holzinger, A. (). Jasplakinolide an actin-specific reagent that promotes actin polymerization. Methods in Molecular Biology, 161, 109-120. Bubb, M., Spector, I., Beyer, B., & Fosen, K. (2000). Effects of Jasplakinolide on the kinetics of actin polymerization. The Journal of Biological Chemistry, 275, 5163-5170.
[14] Peng, G., Wilson, S., & Weiner, O. (2011) A pharmacological cocktail for arresting actin dynamics in living cells. The American society for cell biology.22, 3986-3994.
[15] Stehn, J., Haass, N., Bonello,T., Desouza,M., Kottyan,G., Treutlein, H., Zeng, J., Nascimento, P., Sequeira, N., Butler, T., Allanson, M., Fath, T., Hill, T., McCluskey, A., Schevzov, G., Palmer, S., Hardeman, E., Winlaw, D., Reeve, V., Dixon, I., Weninger, W., Cripe, T., & Peter W. Gunning. (2013). A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Therapeutics, Targets, and Chemical Biology. 73,16.
[16] Goulding, D., Record, J., Malinova, D., Thrasher, A. (2013) Actin cytoskeletal defects in immunodeficiency. Immunol Rev. 256, 282-299.
[17] Vavylonis, D., Yang, Q., & O’Shaughnessy, B. (2005). Actin polymerization kinetics, cap structure, and fluctuations. Proceedings of the National Academy of Sciences of the United States of America. 102, 24.