Talk:20.109(S14):Module 2
Part 1: Transfection of cells with plasmid reporters
All manipulations are to be done with sterile technique in the TC facility.
Timing is important for this experiment, so calculate all dilutions and be sure of all manipulations before you begin.
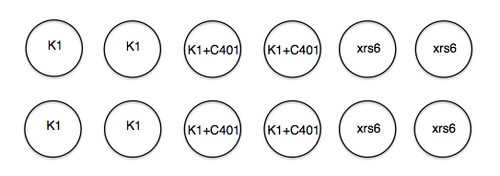
In anticipation of your lipofection experiment, the teaching faculty plated 1x105 K1 and 1.2x105 xrs6 cells per well in a 24-well dish at ~ 11 am yesterday. Half of the K1 cells then received 5 μM of DNA-PK inhibitor "C401" at ~ 5 pm yesterday. A plating schematic for your experiment is shown below. Note carefully which cells are which, including those that have been pre-treated with inhibitor.
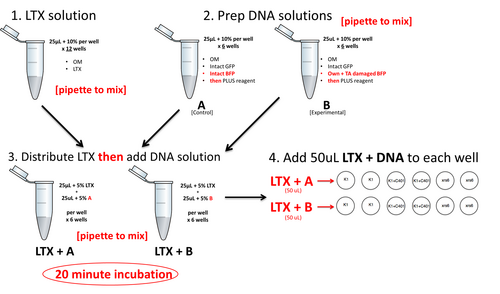
Below is a preview of the lipofection workflow. Do NOT begin these steps until you have completed the calculations further below. At each step, be sure to pipet to mix.
- Pre-label all of the eppendorfs that you will need.
- Prepare as much LTX diluted in OptiMEM (hereafter OM) as needed for all 12 transfections, plus 10%.
- The diluted LTX can sit at room temperature during the next two steps.
- Prepare the DNA mixtures – one control (A) and one experimental (B) – diluted in OM, enough for 6 transfections plus 10%, each.
- Add PLUS reagent to the DNA/OM mixtures.
- Distribute the LTX/OM to fresh eppendorf tubes, one for A and one for B. Here include 5% excess.
- For example, if 50 μL were required, you would add 52.5 instead.
- Then add the DNA/OM/PLUS (also 5% excess) to the LTX/OM.
- Incubate for 20 min at room temperature.
- Add 50 μL of the appropriate LTX/DNA/PLUS/OM mixture to each well. Read through the tips below before proceeding.
- Add the LTX/DNA/PLUS/OM drop-by-drop while making a circle with your pipet in the well, and then immediately swirl the plate (circularly) two times.
- Change tips between every addition.
- After distributing to all 12 wells, do a final mixing step for the whole plate, squeaky style: a few each of horizontal and vertical pushes.
The single-well basis reaction for each of the two DNA mixtures is shown below. Scale each one up to enough for 6 wells worth plus excess, carefully following the workflow above – for example, note that not everything is mixed together at once.
Due to a mistake in planning by the teaching faculty, none of you will have enough DNA to do this experiment with only your own stocks. (Ah, the trials of running a new module!) We recommend that you mix 500 ng of your own DNA with the additional (1000 ng + 150 ng) DNA that you will need for 6 reactions plus excess. You are also welcome to exclusively use the TA-prepared DNA.
| Component | [Stock] ng/μL | Amount per control well (A) | Amount per experimental (B) |
|---|---|---|---|
| LTX (μL) | N/A | 1 | 1 |
| OM for LTX (μL) | N/A | to total 25 μL per well | to total 25 μL per well |
| intact pMax-GFP (ng) | 455.3 | 250 | 250 |
| intact pMax-BFP (ng) | 521.4 | 250 | 0 |
| damaged pMax-BFP-MCS (ng) | unique | 0 | 250 |
| extra cut DNA from Su (ng) | 50 ng/μL | 0 | to supplement 250 above |
| PLUS (μL) | N/A | 0.5 | 0.5 |
| OM for DNA/PLUS (μL) | N/A | to total 25 μL per well | to total 25 μL per well |