TMPRSS2 Lab Notebook
Purpose
TMPRSS2 primes the SARS-CoV-2 spike protein by cleavage of S1/S2 sites which is necessary for virus-cell membrane fusion and cell entry. The purpose of this study is to examine the role of TMPRSS2 polymorphisms and potential impacts they may contribute to SARS-CoV-2.
Combined Method and Results
- Performed a literature search of possible TMPRSS2 SNPs and compiled data into a table
- Link to SNP Table
- This includes individual papers and review papers
- Examine databases for information and data on relevant SNPs
Regulation of TMPRSS2
- TMPRSS2 is mainly expressed in the luminal cells of the prostate epithelium, positively regulated by androgens
- Enhancer located 13 kb upstream of the transcription start site crucial for androgen regulation (Clinckemalie et. al 2013)
- Chromatin looping of E1 and E2 enhancers with PRCAT38 (prostate cancer transcript) and TMPRSS2 promoters (Chen et.al)
- L formation and enhancer activity were mediated by AR/FOXA1 binding and the activity of acetyltransferase p300 (Chen et.al)
- TMPRSS2 has no overt phenotype and mice KO of TMPRSS2 shows no apparent problems, indicate that there might be some functional redundancy with other type II transmembrane serine protease family (TTSP) members. (Thunders and Delahunt 2020)
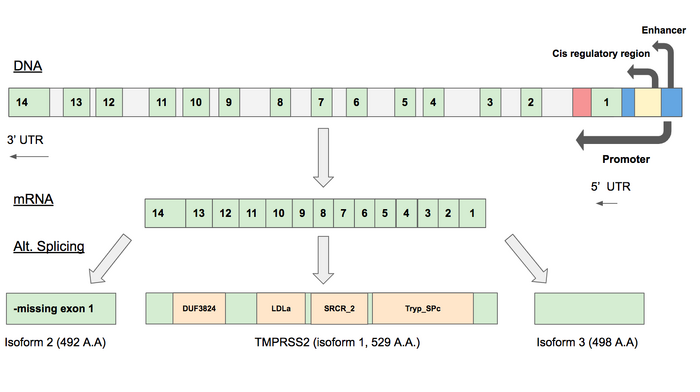
Gene Map
- We created a gene map using Google Slides. TMPRSS2 Gene Diagram
- Text boxes and arrows were used to display regulatory regions and exons.
- Information about exons and regulatory regions were taken from the NCBI gene viewer.
- Isoforms were added to the map.
Isoforms
- TMPRSS2 was searched on Uniprot (UniProtKB - O15393)
- Two isoforms are shown under sequences
- Sequences were compared by clicking the Align button (Alignment)
- Isoform 1 is shown to be lacking the first 37 A.A.
- TMPRSS2 was also searched on NCBI Gene (Gene ID: 7113)
TMPRSS2 Summary
- Full Name: Transmembrane Serine Protease 2
- Chromosome Region: 21q22.3 Location: 41,464,305-41,508,158
- Exons: 14
- Promoter Location: 41,507,255-41,508,648
- Regulatory Regions: 41,508,250-41,508,276
- Enhancers:41,508,128-41,509,135
- Terminator: location unknown
- Has 3 isoforms as a result of alternative splicing
- Isoform 1 is believed to be the most relevant due to its expression in viral target cells.
- DUF3824: Domain of unknown function (44-91 bp)
- LDLa: “Low Density Lipoprotein Receptor Class A” domain: a cysteine-rich repeat that plays a central role in mammalian cholesterol metabolism (150-185 bp)
- SRCR_2: "Scavanger Receptor Cysteine Rich Domain"(190-283 bp)
- Tryp_SPc: “Trypsin-like serine protease”: active site found within this region (293-524 bp)
- Isoform 2 is a 492 amino acid protein that is not yet characterized.
- Isoform 3 is a 498 amino acid protein that is not yet characterized.
ClinVar SNPs
- Relevant snps in the TMPRSS2 gene were researched on ClinVar with the search query "TMPRSS2"
- SNPS were selected that were dependent on the TMPRSS2 gene only.
- Synonymous: rs142750000, rs2298658, rs141788162, rs61735789, rs199824558
- Missense: rs61735793, rs201679623, rs61735790
- Frameshift: rs193920966
Previous Research on TMPRSS2 Structure
- Ravikanth V. et al. (2020) Genetic variants in TMPRSS2 and Structure of SARS-CoV-2 spike glycoprotein and TMPRSS2 complex. bioRxiv. doi:10.1101/2020.06.30.179663
- Alessia D. et al. (2020) Structure, function and variants analysis of the androgen-regulated TMPRSS2, a drug target candidate for COVID-19 infection. bioRxiv. doi:10.1101/2020.05.26.116608
- Hussain, M., Jabeen, N., Amanullah, A., Baig, A. A., Aziz, B., Shabbir, S., ... & Uddin, N. (2020). Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions. AIMS microbiology, 6(3), 350.
Structure
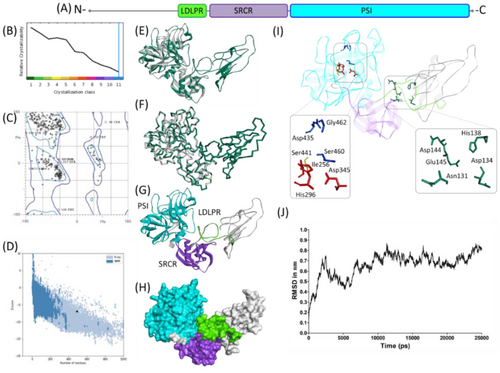
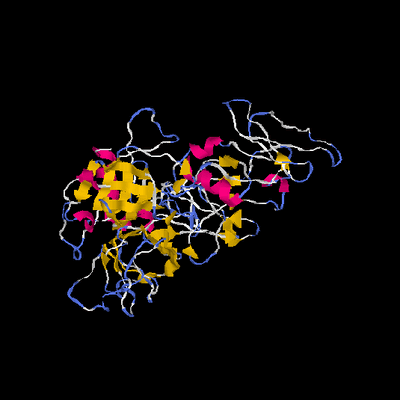
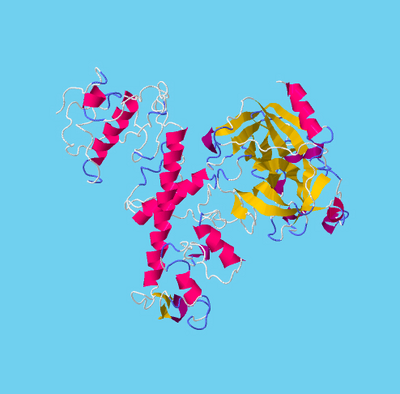
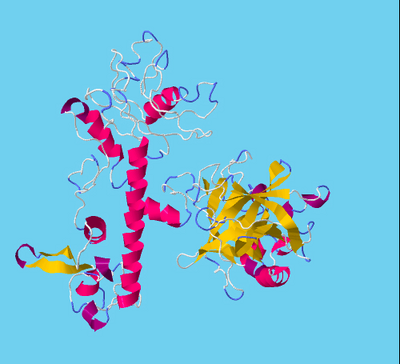
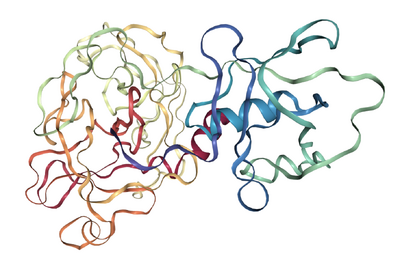
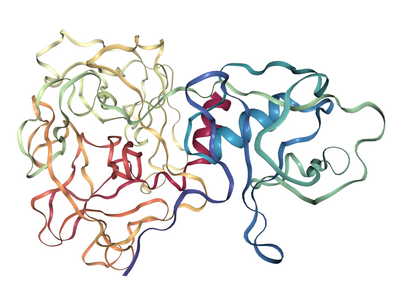
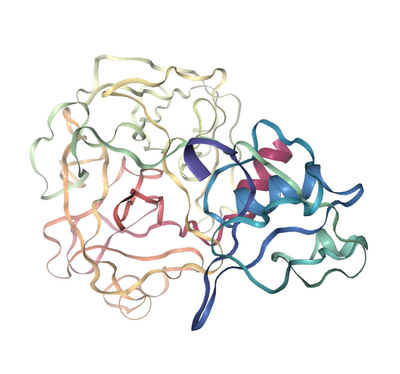
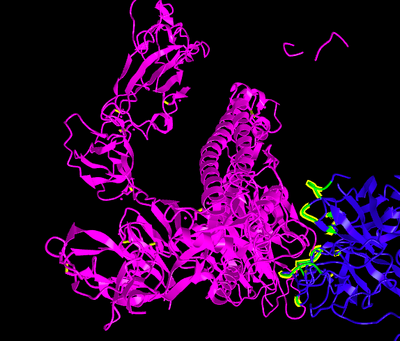
Figure 1. Molecular structure of TMPRSS2. (A) Scaled schematic representation of the functional domains of TMPRSS2 protein (B) XtalPred analysis where blue line represents the least probability of the crystallization (C) Ramachandran plot of the model (D) ProSA analysis of the model where the Z score of the model is indicated by black dot, whereas Z scores of resolved structures are shown with dark blue (NMR) and light blue (X-ray) shades. Superimposition of the full length TMPRSS2 model with template in (E) ribbon (F) Cα backbone conformations. Ribbon conformation (G) and surface topology (H) of TMPRSS2 structure where domains are coloured differently and labelled at the corresponding positions. (I) Functionally important residues are shown in green, blue, red and yellow sticks representing calcium binding sites, substrate binding sites, catalytic sites and proteolytic cleavage site, respectively. (J) Molecular dynamic simulation of TMPRSS2 model showing reasonable stability of the molecule after 10000 picoseconds of the simulation run.
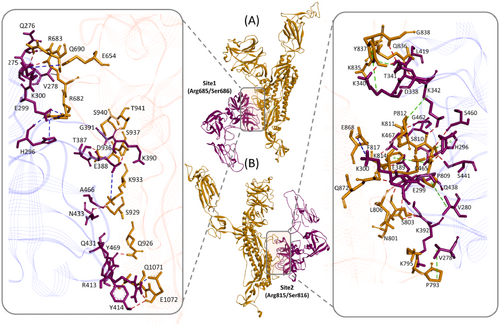
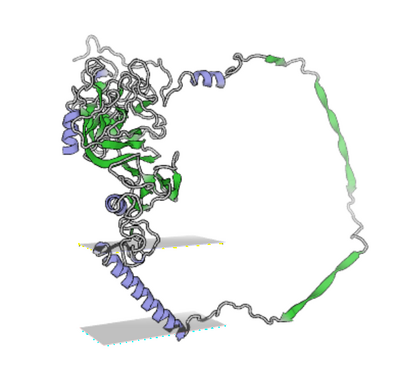
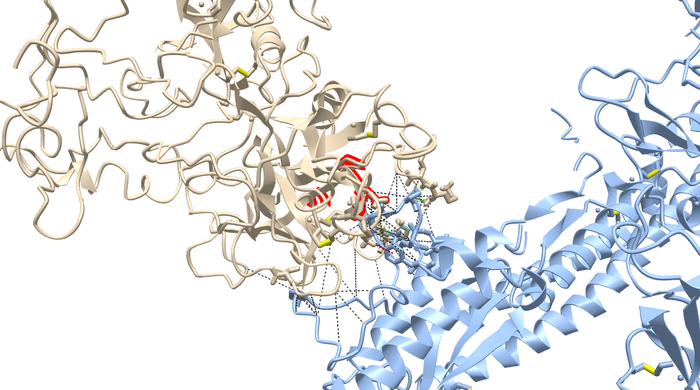
Figure 2. TMPRSS2 and SARS-CoV-2 spike protein Molecular Complex. Ribbon diagram of complexes between TMPRSS2 (magenta) and SARS-CoV-2 spike protein (gold) for (A) site1 (Arg685/Ser686) and (B) site2 (Arg815/Ser816), residues of TMPRSS2 (magenta sticks) and spike protein (gold sticks) involved in the intermolecular interactions are shown in the respective boxes. PDB files of the complexes are made available in supplementary materials Model and Complexes.
- I-TASSER is a protein modeling resource created by Zhang Lab. Uses threading to predict protein secondary and 3D models
- submit protein sequence in FASTA format
- program predicts secondary structure
- predicts if hydrophobic or hydrophilic residues
- Predicts 3D models
- takes about 20 to 60 hours to complete, need to register with the Zhang Lab.
- TMPRSS2 and TMPRSS4 FAFSA sequence were inputted into database.
- TMPRSS2 model in I-TASSER
- TMPRSS4 model in I-TASSER
- Protein structure homology-modelling
- Has TMPRSS2 modeled already for isoform 1 ID:O15393
Swiss-Model Run for TMPRSS2 & rs12329760
Input FASTA data for TMPRSS2 with and without rs12339760 mutation (V160 --> M)
- FASTA format for TMPRSS2 was obtained from Uniprot:O15393
>sp|O15393-2|TMPS2_HUMAN Isoform 2 of Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSGSPPAIGPYYENHGYQPE NPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKK ALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGE DENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGIVDD SGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNSSRQSRIVGGESAL PGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEKPLNNPWHWTAFAGILRQSFMFYG AGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISG WGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGG PLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG
- rs12329760 mutation was made on the A.A. sequence (V160 --> M)
>sp|O15393-2|TMPS2_HUMAN Isoform 2 of Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSGSPPAIGPYYENHGYQPE NPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKK ALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGE DENRCVRLYGPNFILQMYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGIVDD SGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNSSRQSRIVGGESAL PGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEKPLNNPWHWTAFAGILRQSFMFYG AGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISG WGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGG PLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG
- TMPRSS2 run: Job ID:KtuPmY
- TMPRSS2 w/ rs12329760 run: Job ID:5UeD3B
- Software from BonvinLab - models interaction between two molecular structures and how they fit together.
- Must register to the website, submit a new job, fill out required field (job name, type of structures), click Next and generate results.
- ClusPro is another website to look at protein-protein docking, needs PDB and chains
- SARS-Cov-2 S protein was inputted into HADDOCK using the ID: 7DK3, chain A, Protein-Protein Ligand Docking
- TMPRSS2 3D structure was inputted into HADDOCK using the PDB file obtained from I-TASSER, Protein-Protein Ligand Docking
- Known cleavage site of S2' region put in for "Active Residues" of SARS-CoV-2 Arg815-Ser816
- "Active residues of TMPRSS2": (from Hussain 2020).
- His296, Asp345, Ser441 (catalytic triad)
- Asp435, Ser460, Gly462 (substrate binding site)
- RaptorX is a protein structure prediction server developed by the Xu group. When a sequence is input, RaptorX can predict secondary and tertiary protein structures, contacts, solvent accessibility, disordered regions and binding sites.
- To submit a job to RaptorX users should
- Register with email for quick retrieval of results
- Input a protein sequence or upload a FASTA file. Wait time is 2-3 days.
- Retrieve results with job ID, email, or sequence
- Results will include a predicted contact map, a contact result file, and five predicted 3D models assisted by the predicted contacts
RaptorX prediction of TMPRSS2 (Isoform 2) and rs12329760
- FASTA format for TMPRSS2 was obtained from Uniprot: O15393
>sp|O15393-2|TMPS2_HUMAN Isoform 2 of Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSGSPPAIGPYYENHGYQPE NPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKK ALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGE DENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGIVDD SGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNSSRQSRIVGGESAL PGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEKPLNNPWHWTAFAGILRQSFMFYG AGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISG WGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGG PLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG
- rs12329760 mutation was made on the A.A. sequence (V160 --> M)
>sp|O15393-2|TMPS2_HUMAN Isoform 2 of Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSGSPPAIGPYYENHGYQPE NPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKK ALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGE DENRCVRLYGPNFILQMYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGIVDD SGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNSSRQSRIVGGESAL PGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEKPLNNPWHWTAFAGILRQSFMFYG AGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISG WGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGG PLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG
- FASTA sequences were separately input into RaptorX
- Predicted contact and 3D models were generated
- TMPRSS2 JobID 16764609
- rs12329760 JobID 10704949
- HHpred can be utilized for detecting remote protein homology and structure prediction, including secondary and tertiary structure.
- Involves information from databases including PDB, SCOP, Pfam, SMART, COGs, and CDD.
- To visualize a protein structure, users can:
- Input the protein sequence in A3M/FASTA/CLUSTAL/STOCKHOLM format into the Input field and select submit.
- Using the results generated, select the templates you would like to visualize and then select Create Model Using Selection.
- This will generate a PIR file, which can be pasted into the MODDELLER software under 3ary structure.
- To download and run the MODDELLER software, users need to register for a license key
- Inputing this license key into Custom Job ID and clicking Submit will generate results.
HHpred for TMPRSS2 in the absence and presence of rs12329760
- FASTA format for TMPRSS2 was obtained from Uniprot:O15393
>sp|O15393-2|TMPS2_HUMAN Isoform 2 of Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSGSPPAIGPYYENHGYQPE NPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKK ALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGE DENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGIVDD SGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNSSRQSRIVGGESAL PGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEKPLNNPWHWTAFAGILRQSFMFYG AGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISG WGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGG PLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG
- rs12329760 mutation was made on the A.A. sequence (V160 --> M)
>sp|O15393-2|TMPS2_HUMAN Isoform 2 of Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 MPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSGSPPAIGPYYENHGYQPE NPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKK ALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGE DENRCVRLYGPNFILQMYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGIVDD SGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNSSRQSRIVGGESAL PGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEKPLNNPWHWTAFAGILRQSFMFYG AGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISG WGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGG PLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG
- FASTA sequences were imputed into HHpred in separate runs and submitted.
- TMPRSS2: Job ID:5623875
- TMPRSS2 rs12329760: Job ID: 5623875_2
- The protein templates generated from these runs were visualized using MODDELLER.
- TMPRSS2: Job ID: 8753897
- TMPRSS2 rs1239760: Job ID: 8753897
- HHpred shows no effect on protein structure on rs12329760. Both proteins did not have A.A. 160 included in their structure
- PredMP is a de novo prediction and visualization of membrane proteins
- TMPRSS2 is a transmembrane serine protease
- FASTA sequence for TMPRSS2 was imputted into the server
- TMPRSS2: Job ID: 20111411494111
- It gave 5 predicted models of TMPRSS2
- First de novo prediction model of TMPRSS2
SNPs of interest
- Possible SNPs of interest include ones listed in ClinVar
- Synonymous: rs142750000, rs2298658, rs141788162, rs61735789, rs199824558
- Missense: rs61735793, rs201679623, rs61735790
- Frameshift: rs193920966
- SNP rs12329760 which has been seen in multiple papers that is a V --> M mutation
- SNP rs2070788 and rs383510 has been cited in multiple papers as conferring higher risk for severe influenza A virus infection (G-->A)
- SNP rs456142 and rs462574 are 3' UTR variants that are located in mRNA target sites.
- SNP rs456298 is a 3' UTR variant that has a different frequency for Asian populations and may be correlated with immune response to rubella vaccine.
- SNP rs75603675 has been cited in multiple papers that might be associated with SARS-CoV-2 entry
- SNP rs139010197 is a TMPRSS11a missense variant and has been cited to may increase infection risk [1]
- SNP rs977728 is a TMPRSS11a initiator codon missense variant and may increase infection risk [2]
- SNP rs353163 is a TMPRSS11a missense variant that has been cited by multiple papers to increase risk to esophageal cancer.
- Russo et. al stated that the SNP rs1475908, whose alternative allele (A) is associated with low TMPRSS2 expression, and the two variants rs74659079 (allele T) and rs2838057 (allele A), both associated with high TMPRSS2 expression. Interestingly, the eQTL rs1475908 shows the highest AF among EAS (A:0.38) and EUR (A:0.35) and the lowest frequency among Latinos (0.17) These findings agree with a previous study that demonstrated the association of two high TMPRSS2 expression-variants, rs2070788 (allele G) and rs383510 (allele T), with increased susceptibility to the influenza virus infection A (H7N9)
SNP Population Frequencies
Allele frequencies for each SNP from above was found by searching the SNP on dbSNP and looking under “Frequency”.
- For consistency across SNPs, Allele Frequency Aggregator ALFA was used for the collection of data.
Frameshift
- Population frequencies not available for rs193920966
Intron Variants
| Population | European | African | Asian | Latin American 2 | Total |
|---|---|---|---|---|---|
| rs2070788 | G=0.461512 A=0.538488 | G=0.3100 A=0.6900 | G=0.325 A=0.675 | G=0.5271 A=0.4729 | G=0.459889 A=0.540111 |
| rs383510 | T=0.4817 C=0.5183 | T=0.33 C=0.67 | T=0.5 C=0.5 | T=0 C=0 | T=0.4771 C=0.5229 |
Missense
| Population | European | African | Asian | Latin American 2 | Total |
|---|---|---|---|---|---|
| rs61735793 | G=0.9901 A=0.0099 | G=0.999 A=.001 | G=1.000 A=0.000 | G=1.00 A=0.00 | G=0.99025 A=0.00975 |
| rs201679623 | A=0.9999 C=0.00001 | A=1.000 C=0.000 | A=1.000 C=0.000 | A=1.000 C=0.000 | A=1.000 C=0.000 |
| rs61735790 | T=0.99996 C=0.00004 | T=0.993 C=0.007 | T=1.000 C=0.000 | T=1.00 C=0.00 | T=0.99989 C=0.00011 |
| rs12329760 | C=0.777704 T=0.2222 | C=0.7093 t=0.2907 | C=0.620 T=0.380 | C=0.8536 T=0.1464 | C=0.777066 T=0.2229 |
| rs75603675 | C=0.6017 A=0.3983 | C=0.68 A=0.32 | C=1.0 A=0.0 | C=0 A=0 | C=0.6059 A=0.3941 |
| rs139010197 | T=0.97531 C=0.02469 | T=0.982 C=0.018 | T=1.000 C=0.000 | T=1.00 C=0.00 | T=0.97552 C=0.02448 |
| rs977728 | C=0.823458 T=0.176542 | C=0.8676 T=0.1324 | C=0.824 T=0.176 | C=0.65 T=0.35 | C=0.823073 T=0.176927 |
| rs353163 | T=0.33089 C=0.66911 | T=0.1788 C=0.8212 | T=0.173 C=0.827 | T=0.4638 C=0.5362 | T=0.329175 C=0.670825 |
3' UTR Variant
| Population | European | African | Asian | Latin American 2 | Total |
|---|---|---|---|---|---|
| rs456142 | T=0.1481 C=0.8300 | T=0.406 C=0.594 | T=0.67 C=0.33 | T=0.33 C=0.67 | T=0.1700 C=0.8300 |
| rs462574 | A=0.02352 G=0.97648 | A=0.1818 G=0.8182 | A=0.516 G=0.484 | A=0.2661 G=0.7339 | A=0.05383 G=0.94617 |
| rs456298 | T=0.1515 A=0.8485 | T=0.43 A=0.57 | T=0.8 A=0.2 | T=0 A=0 | T=0.1654 A=0.8346 |
Synonymous
| Population | European | African | Asian | Latin American 2 | Total |
|---|---|---|---|---|---|
| rs142750000 | C=0.99555 T=0.00445 | C=0.999 T=0.001 | C=1.000 T=0.000 | C=1.00 T=0.00 | C=0.99574 T=0.00426 |
| rs2298658 | C=1.0000 T=0.0000 | C=1.00 T=1.00 | C=1.0 T=0.0 | C=0 T=0 | C=1.000 T=0.000 |
| rs141788162 | G=0.9951 A=0.0049 | G=0.988 A=0.012 | G=0.98 A=0.02 | no data | G=0.99436 A=0.00564 |
| rs61735789 | G=0.98298 A=0.01702 | G=0.987 A=0.013 | G=1.00 A=0.00 | no data | G=0.98327 A=0.01673 |
| rs199824558 | G=0.9980 A=0.0020 | G=0.999 A=0.001 | G=1.00 A=0.00 | no data | G=0.99821 A=0.00179 |
- PolyPhen2 predicts whether certain amino acid substitutions will be damaging or non-damaging to the protein
- TMPRSS2 SNPs of interest were submitted into queries to see if they are expected to be damaging
- Probably Damaging: rs12329760
TMPRSS2 SNP Predictions
- dbSNP ID was entered
- SIFT score and SIFT predictions were recorded
- SIFT scores range from 0 to 1. The amino acid substitution is considered damaging if the score is less than 0.05 and is tolerated if the score is greater than 0.05.
- PolyPhen-2 scores and predictions were recorded
- Polyphen-2 scores range from 0 to 1. The amino acid substitution is considered benign if the score is less than 0.05, possibly damaging if the score is between 0.5 and 0.9, and probably damaging if the score is between 0.9 and 1.
| rs Number | SIFT score | SIFT prediction | PolyPhen-2 score | PolyPhen-2 prediction |
|---|---|---|---|---|
| rs61735793 | 0.238 | tolerated | 0.015 | Benign |
| rs75603675 G8V | 0.201 | tolerated | 0.167 | Benign |
| rs61735790 | 0.231 | tolerated | 0.033 | Benign |
| rs12329760 | 0.009 | deleterious | 0.937 | Probably Damaging |
| rs200291871 | 0.817 | Tolerated | 0.011 | Benign |
| rs61735791 | 0.199 | Tolerated | 0.029 | Benign |
| rs148125094 | 0.171 | Tolerated | 0.098 | Benign |
| rs114363287 | 0.383 | Tolerated | 0.109 | Benign |
| rs147711290 L128G | Not Found | - | 0.920 | Probably Damaging |
| rs147711290 L91P | 0.005 | Deleterious | 1.000 | Probably Damaging |
| rs147711290 L91R | Not Found | - | Not Found | - |
| rs150554820 | 0.004 | Deleterious | 0.549 | Possibly Damaging |
| rs61735796 | 0.34 | Tolerated | 0.017 | Benign |
| rs138651919 | 0.021 | Deleterious | 0.833 | Possibly Damaging |
| rs61735795 | 0.551 | Tolerated | 0.086 | Benign |
| rs142446494 | 0.015 | Deleterious | 0.294 | Benign |
| rs201093031 | 1 | Tolerated | 0.00 | Benign |
| rs768173297 | Not Found | - | 0.131 | Benign |
TMPRSS4
- TMPRSS4 may serve a similar function in viral entry as TMPRSS2, as it also codes for a membrane bound serine protease that was observed to recognize the SARS-CoV-2 spike protein and help facilitate its entry into cells.
- SNPS of interest: rs142842357 (ClinVar),
- High expression (>90%) in tissues such as the nasal cavity, esophagus, bronchus epithelium, colon, intestine, and oral cavity. Source
- TMPRSS4 snps were filtered on the NCBI dnSNP database by frequency in populations (MAF 0.05-0.1)
- 84 SNPs were found as of January 19th 2021
- None of the SNPs had citations or Clinvar significance.
TMPRSS4 and SARS-CoV-2
- TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes-Zang et al.
- TMPRSS4 increases SARS-CoV-2 infectivity in intestinal epithelial cells.
- When TMPRSS4 was expressed with ACE2 in HEK293 cells, there was an increase in viral RNA levels.
- TMPRSS4 knockout led to a 4x reduction in VSV-SARS-CoV-2 replication in human enteroid cells, much greater than seen in TMPRSS2 knockout cells.
- Higher TMPRSS4 levels are correlated with COVID-19 infection.
Structural Model of TMPRSS4
- 3D Visualization of TMPRSS4 was generated by importing the FASTA sequence into HHPred's MODELLER software.
- The FASTA sequence of TMPRSS4 was taken from Uniprot: Q9NRS4
>sp|Q9NRS4|TMPS4_HUMAN Transmembrane protease serine 4 OS=Homo sapiens OX=9606 GN=TMPRSS4 PE=1 SV=2 MLQDPDSDQPLNSLDVKPLRKPRIPMETFRKVGIPIIIALLSLASIIIVVVLIKVILDKY YFLCGQPLHFIPRKQLCDGELDCPLGEDEEHCVKSFPEGPAVAVRLSKDRSTLQVLDSAT GNWFSACFDNFTEALAETACRQMGYSSKPTFRAVEIGPDQDLDVVEITENSQELRMRNSS GPCLSGSLVSLHCLACGKSLKTPRVVGVEEASVDSWPWQVSIQYDKQHVCGGSILDPHWV LTAAHCFRKHTDVFNWKVRAGSDKLGSFPSLAVAKIIIIEFNPMYPKDNDIALMKLQFPL TFSGTVRPICLPFFDEELTPATPLWIIGWGFTKQNGGKMSDILLQASVQVIDSTRCNADD AYQGEVTEKMMCAGIPEGGVDTCQGDSGGPLMYQSDQWHVVGIVSWGYGCGGPSTPGVYT KVSAYLNWIYNVWKAEL
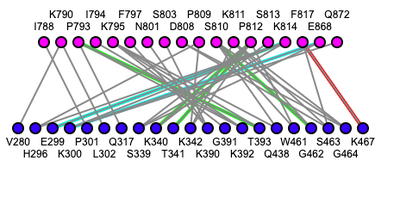
- Interaction between TMPRSS2 and SARS-CoV-2 highlighted in yellow and green.
- Upload docked PDB file into iC3nD, click Analysis, View Sequence, Interactions, go to the Details, then highlight the relevant interactions in the bottom bar. Interactions will be highlighted in the model view.
- Interaction Network was shown by clicking Analysis, H-bonds and Interactions, For the first set pick Structure A and then for the second set Structure B, click Interaction Network to generate the network image.
TMPRSS2 missense SNPs in FASTA format
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs61735793 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGIVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs201679623 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYDPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs61735790 MALNSGSPPAIGPYYENRGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs12329760 MALNSGSPPAIGPYYENRGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQMYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs61735791 MALNSGSPPAIGPYYENHGYQPENPYPTQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs148125094 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYIYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs114363287 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSRTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs147711290 (L91R) MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTRGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs147711290 (L91P) MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTPGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs147711290 (L91Q) MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTQGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs150554820 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSIMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs61735796 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGKSALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs138651919 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHLAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs61735795 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQSEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs142446494 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHMCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs201093031 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVAPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
>sp|O15393|TMPS2_HUMAN Transmembrane protease serine 2 OS=Homo sapiens OX=9606 GN=TMPRSS2 PE=1 SV=3 rs768173297 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK PLNNPWHWMAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF TDWIYRQMRADG
TMPRSS2 Multiple Sequence Alignment
CLUSTAL FORMAT: MUSCLE (3.8) multiple sequence alignment
s_147Q MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_147P MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_147R MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_63 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s.O.TMPS2 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_65 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_14812509 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_61 MALNSGSPPAIGPYYENHGYQPENPYPTQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_12329760 MALNSGSPPAIGPYYENRGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_60 MALNSGSPPAIGPYYENRGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_2679623 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYDPSPVPQYAPRVLTQA
s_11436328 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_15055482 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_66 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_13865191 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHLAQYYPSPVPQYAPRVLTQA
s_2093031 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVAPTVYEVHPAQYYPSPVPQYAPRVLTQA
s_76817329 MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
s MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVPQYAPRVLTQA
*****************.*********:****.******* *** ***************
s_147Q SNPVVCTQPKSPSGTVCTSKTKKALCITLTQGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_147P SNPVVCTQPKSPSGTVCTSKTKKALCITLTPGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_147R SNPVVCTQPKSPSGTVCTSKTKKALCITLTRGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_63 SNPVVCTQPKSPSGIVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s.O.TMPS2 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_65 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_14812509 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_61 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_12329760 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_60 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_2679623 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_11436328 SNPVVCTQPKSPSRTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_15055482 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_66 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_13865191 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_2093031 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s_76817329 SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
s SNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAALAAGLLWKFMGSKCSNSGIEC
************* *************** *****************************
s_147Q DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_147P DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_147R DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_63 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s.O.TMPS2 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_65 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_14812509 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_61 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_12329760 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQMYSSQRKSWHPVCQDDWNENY
s_60 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_2679623 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_11436328 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_15055482 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_66 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_13865191 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_2093031 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s_76817329 DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
s DSSGTCINPSNWCDGVSHCPGGEDENRCVRLYGPNFILQVYSSQRKSWHPVCQDDWNENY
***************************************:********************
s_147Q GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_147P GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_147R GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_63 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s.O.TMPS2 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_65 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_14812509 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_61 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_12329760 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_60 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_2679623 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_11436328 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_15055482 GRAACRDMGYKNNFYSSQGIVDDSGSTSIMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_66 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_13865191 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_2093031 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s_76817329 GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
s GRAACRDMGYKNNFYSSQGIVDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLR
****************************:*******************************
s_147Q CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_147P CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_147R CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_63 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s.O.TMPS2 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHMCGGSIITPEWIVTAAHCVEK
s_65 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_14812509 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_61 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_12329760 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_60 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_2679623 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_11436328 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_15055482 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_66 CIACGVNLNSSRQSRIVGGKSALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_13865191 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_2093031 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s_76817329 CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
s CIACGVNLNSSRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK
*******************:*******************:********************
s_147Q PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_147P PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_147R PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_63 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s.O.TMPS2 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_65 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_14812509 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_61 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_12329760 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_60 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_2679623 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_11436328 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_15055482 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_66 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_13865191 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_2093031 PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s_76817329 PLNNPWHWMAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
s PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL
******** ***************************************************
s_147Q VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_147P VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_147R VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_63 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s.O.TMPS2 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_65 VKPVCLPNPGMMLQSEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_14812509 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYIYDNLI
s_61 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_12329760 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_60 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_2679623 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_11436328 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_15055482 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_66 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_13865191 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_2093031 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s_76817329 VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
s VKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLI
**************.***************************************:*****
s_147Q TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_147P TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_147R TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_63 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s.O.TMPS2 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_65 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_14812509 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_61 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_12329760 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_60 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_2679623 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_11436328 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_15055482 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_66 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_13865191 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_2093031 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s_76817329 TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
s TPAMICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTSWGSGCAKAYRPGVYGNVMVF
************************************************************
s_147Q TDWIYRQMRADG
s_147P TDWIYRQMRADG
s_147R TDWIYRQMRADG
s_63 TDWIYRQMRADG
s.O.TMPS2 TDWIYRQMRADG
s_65 TDWIYRQMRADG
s_14812509 TDWIYRQMRADG
s_61 TDWIYRQMRADG
s_12329760 TDWIYRQMRADG
s_60 TDWIYRQMRADG
s_2679623 TDWIYRQMRADG
s_11436328 TDWIYRQMRADG
s_15055482 TDWIYRQMRADG
s_66 TDWIYRQMRADG
s_13865191 TDWIYRQMRADG
s_2093031 TDWIYRQMRADG
s_76817329 TDWIYRQMRADG
s TDWIYRQMRADG
************
PredictProtein
PredictProtein is a program that can be used to predict secondary structure, solvent accessibility, transmembrane helices, globular region, and much more.
- FASTA format of TMPRSS2 was inputted into text box
- Results can be seen [Here]
SNPs on TMPRSS2-SARS-CoV-2 Docking Analysis
SNPs below are from most frequent to low frequent:
- V160M- not near interactions but in SRCR conserved domain, predicted to be lethal using PredictProtein heatmap and PolyPhen2 & SIFT
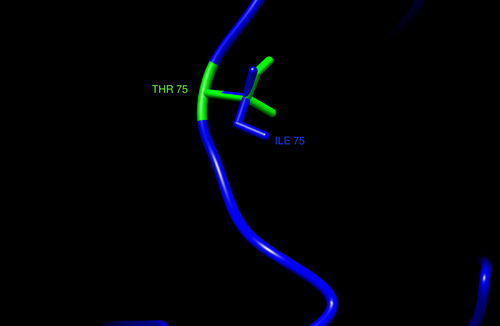
- Thr75Ile- not near interactions
- Ala28Thr- not near interactions
- Val415Ile- close to interaction sites, in the serine protease domain, predicted to have little-no effect on PredictProtein
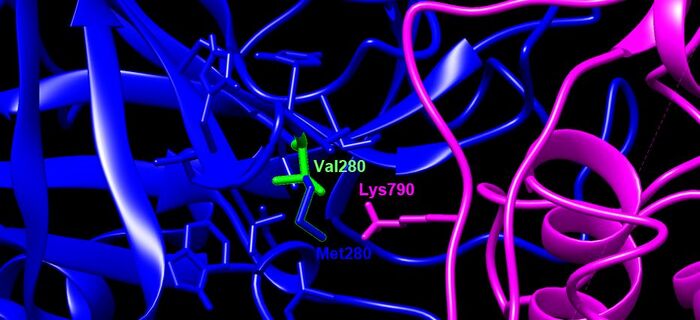
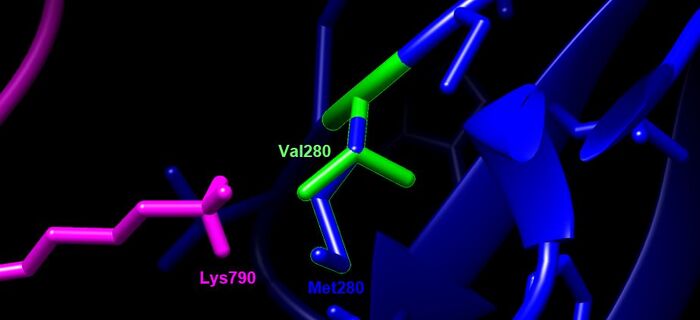
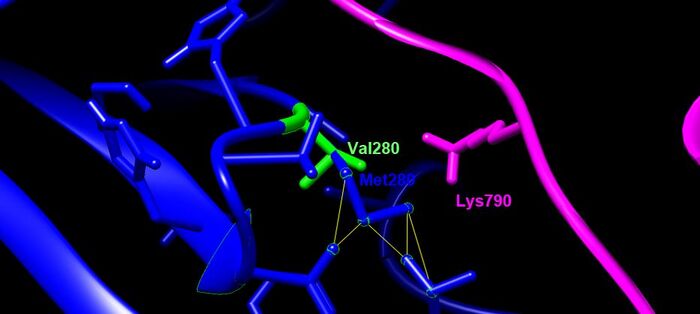
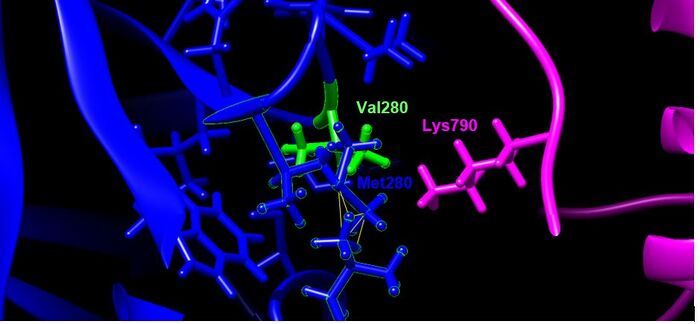
- Val280Met- This is an interaction site. It is located on a beta sheet and interacts with K790 on the Spike Protein (right after fusion peptide). Valine to Methionine are both nonpolar amino acids, but there is a gain of a sulfur atom which could potentially unfavorably bind to nearby amino acids.
- Glu260Lys- somewhat close to interactions with the spike protein, in the serine protease domain. Predicted to be tolerated in SIFT and PolyPhen2.
- Phe209Ile- not near interactions but in SRCR conserved domain, predicted to be deleterious/lethal in PolyPhen2 and SIFT, and a red signal in the PredictProtein heatmap.
- Pro41Leu- not near interactions
- His18Arg- not near interactions
- Thr309Met- This is partially close to several interaction sites (302,301,300). It is located in a beta sheet and predicted have little/no effect in PredictProtein heatmap. (PolyPhen-2 and SIFT could not be determined).
- Pro275Ser- not near interactions
- Val33Ala- not near interactions
- Leu91Gln- not near interactions
- Leu91Pro- not near interactions
- Leu91Arg- not near interactions
- Gly74Arg- not near interactions
- Leu81Arg- not near interactions
Secondary Structure Alignment
- HH=HHPred, SM=SWISS-MODEL, RX=RaptorX, I-T=I-TASSER, AA= amino acid sequence
- H = helix, B = beta strand, - = random coil, space = not modeled
RX -------------------------------------------------- I-T -------------------------------------------------- HH SM AA MALNSGSPPAIGPYYENHGYQPENPYPAQPTVVPTVYEVHPAQYYPSPVP 50 RX -----------------------------HHHHHHHHHHHHHHHHHHHHH I-T -------------------------------------------------- HH SM AA QYAPRVLTQASNPVVCTQPKSPSGTVCTSKTKKALCITLTLGTFLVGAAL 100 RX HHHHHHHH---------BBBHHH-BBB---------------HHHHHH-- I-T ------------------------------------------------BB HH SM -----BB AA AAGLLWKFMGSKCSNSGIECDSSGTCINPSNWCDGVSHCPGGEDENRCVR 150 RX -------------HHH-----------HHHHHHHHHH------------- I-T B------BBBBBHHH-BBBB------HHHHHHHHHHHH----BBBBBBBB HH -BBBBB---BBBB------HHHHHHHHHHHH----BBBBBBBB SM B------BBBBB----BBBB------HHHHHHHHHHHH----BBBBBBBB AA LYGPNFILQVYSSQRKSWHPVCQDDWNENYGRAACRDMGYKNNFYSSQGI 200 RX ----------------------------HHHHHHHHHHHHHHH------- I-T ----------------------------------BBBBBBB--------- HH --------BBBB---HHHH--HHHHBBBB-----BBBBBBB--------- SM HHHH-----BBB---HHH---HHHHBBB------BBBBBBB--------- AA VDDSGSTSFMKLNTSAGNVDIYKKLYHSDACSSKAVVSLRCIACGVNLNS 250 RX ---------BB--------BBBBBB--BBBBBBBBBB--BBBBBHHH--- I-T --------BBB--------BBBBBB--BBBBBBBBBB--BBBB-HHHH-- HH ----------BB-------BBBBBB--BBBBBBBB----BBBB-HHH--H SM -------------------BBBBBB---BBBBBBBBB--BBB--HHHH-H AA SRQSRIVGGESALPGAWPWQVSLHVQNVHVCGGSIITPEWIVTAAHCVEK 300 RX -------BBBBB--------HHHBBBBBBBBBB------------BBBBB I-T ----HHHBBBBB-----------BBB-BBBBBB-------------BBBB HH HH--HHHBBBBB-----------BBB-BBBBBB-------------BBBB SM HH--HHHBBBBB-----------BBB-BBBBBB-------------BBBB AA PLNNPWHWTAFAGILRQSFMFYGAGYQVEKVISHPNYDSKTKNNDIALMK 350 RX ---------------------------BBBBBBB------------BBBB I-T B--------------------------BBBBBB--------------BBB HH ---------------------------BBBB----------------BB- SM B--------------------------BBBBBB--------------BBB AA LQKPLTFNDLVKPVCLPNPGMMLQPEQLCWISGWGATEEKGKTSEVLNAA 400 RX BBBBB-HHHH--HHH--------BBBBB---------HHH--BBBBBBB- I-T BBBBB------------------BBBBB---------------BBBBBB- HH BBBBB-HHHHH------------BBBB----------------BBBBBB- SM BBBBB-HHHH-------------BBBB----------------BBBBB-- AA KVLLIETQRCNSRYVYDNLITPAMICAGFLQGNVDSCQGDSGGPLVTSKN 450 RX -BBBBBBBBBBBHHH------BBBBBHHH-HHHHHHHHHHH I-T -BBBBBBBBBB----------BBBBBHHHHHHHHHHH----- HH -BBBBBBBBBB----------BBBBBHHHHHHHHHHHHHH- SM --BBBBBBBBB----------BBBBBHHHHHHHHHHHHHH AA NIWWLIGDTSWGSGCAKAYRPGVYGNVMVFTDWIYRQMRADG 492
Assessment of TMPRSS2 Model
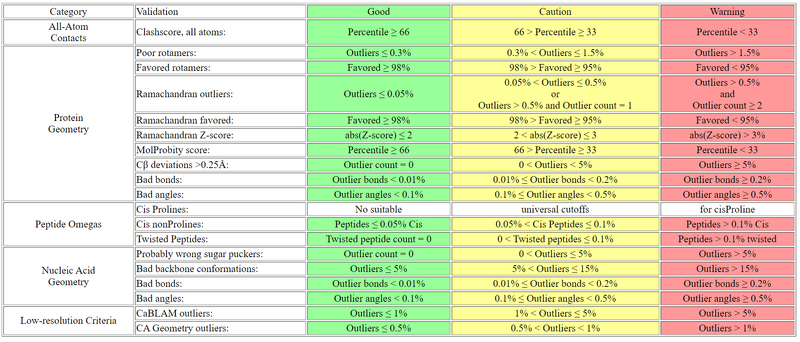
- Ramachandran(phi/psi) plots can be used to understand which secondary structures of proteins are sterically allowed/favored to occur
- Comparing the structures of TMPRSS2 generated to a Ramachandran plot of TMPRSS2 allows us to see if the predicted structure contains elements that would/would not be sterically allowed to occur and therefore judge the accuracy of our structure
- Ramachandran plot generating softwares:
- MolProbity: cited in a similar study and allows for input of any PDB file
- Uppsala Ramachandran Server: generates a Ramachandran plot for structures already deposited in the PDB with specific code.
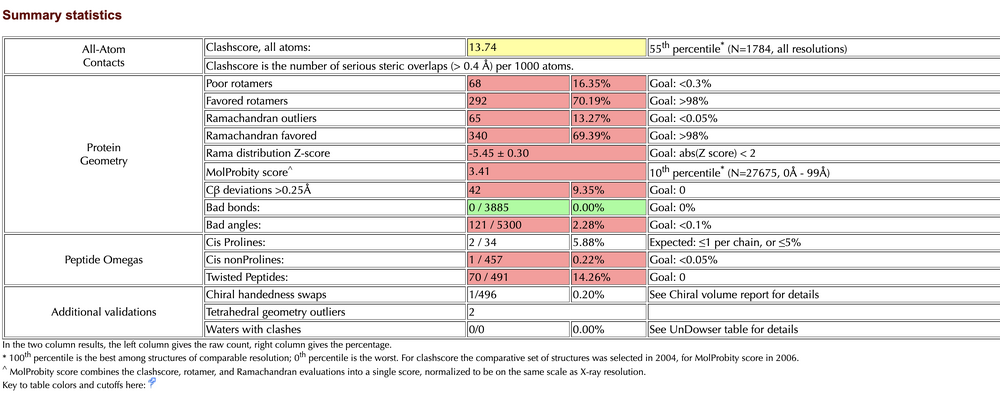
- To generate Ramachandran plots, PDB files of TMPRSS2 from each modelling software (RaptorX, I-Tasser, HHpred, Swiss-Model) was uploaded onto MolProbity by selecting Choose File, uploading the file, then selecting Upload
- After PDB has uploaded, select Analyze geometry without all-atom contacts
- 'RaptorX_TMPRSS2.pdb' was selected and all default outputs were run
Summary Table Cutoffs
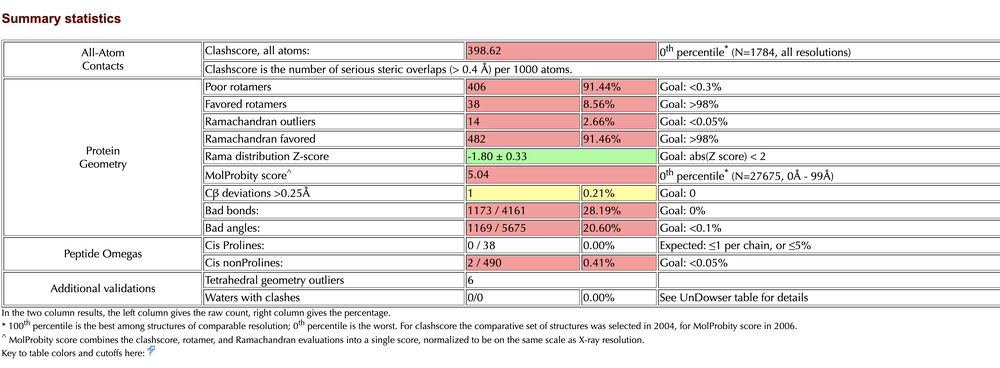
RaptorX
- Summary Statistics of Predicted Structure
- Ramachandran Plot for TMPRSS2 generated by Raptor X
Ramachandran Plot (PDF)
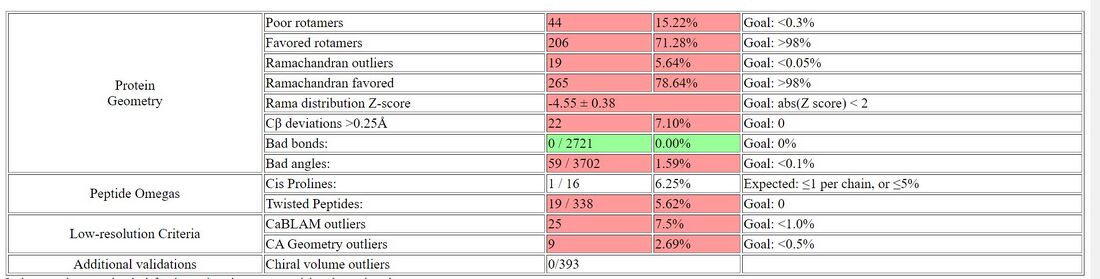
I-TASSER
- Summary Statistics of Predicted Structure
- Ramachandran plot of TMPRSS2 generated by I-TASSER
Ramachandran plot (PDF)
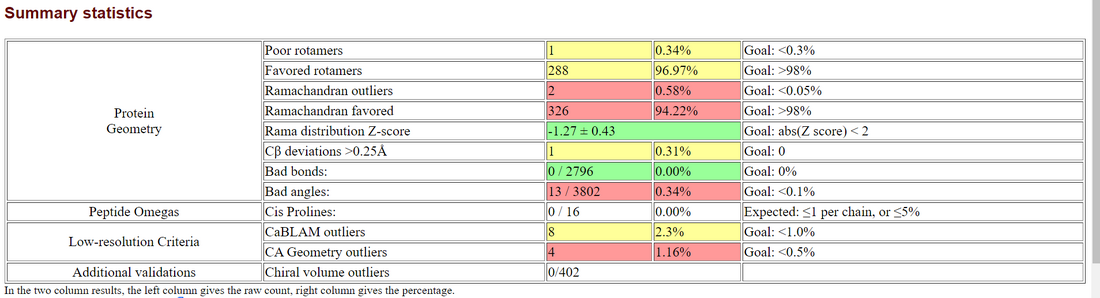
SWISS-MODEL
- Summary statistics of Predicted Structure
- Ramachandran plot of structure generated by SWISS-MODEL
Ramachandran plot (pdf)
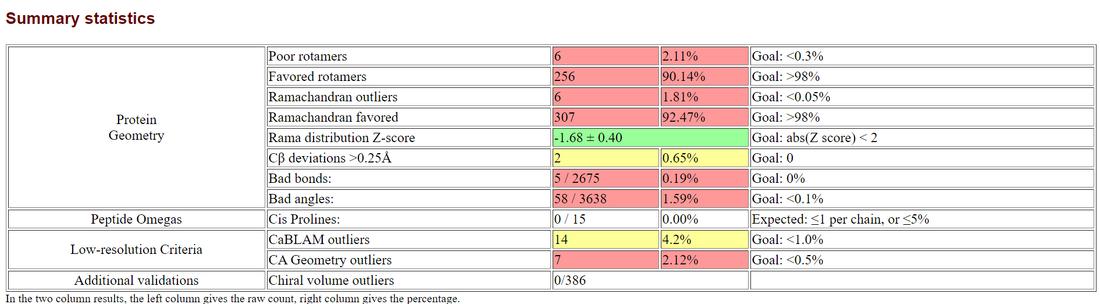
HH-Pred
- Summary Statistics of Predicted Structure
- Ramachandran plot of structure generated by HH-Pred
Ramachandran plot (pdf)
Truncated I-TASSER
- Residues 149-487
- Summary Statistics of Predicted Structure
- Ramachandran plot of structure generated by I-TASSER
Ramachandran plot (pdf)
To Do
- fix secondary structure alignment to take out spaces
- look at Ramachandran plots
- divide number of outliers by number of amino acids modeled
- Raptor X: 14 outliers / 492 = 2.65%
- I-TASSER: 65 outliers / 492 = 13.27%
- SWISS-Model: 2 outliers / 492 = 0.57%
- HHPred: 6 outliers / 492 = 1.81%
- look at iTasser to see if most of the outliers are in the random coil at the beginning of the structure
- Most I-TASSER outliers occur from 8-150 aa
- the shortest model is HH, so look to see what proportion of outliers are in all of them for the length of HH (HHPred models from 158 aa to 491 aa)
- Raptor X: 13 outliers / 332 = 3.92%
- I-TASSER: 18 outliers / 332 = 5.42%
- Swiss-Model: 2 outliers / 332 = 0.60%
- divide number of outliers by number of amino acids modeled
- if it's easy, you could chop off the random coil part of the iTasser model and just run that through the Ramachandran plots
Chimera
How to Mutate a Residue
- Input docked model, select TMPRSS2 chain, select Chain B (if chain B is TMPRSS2)
- To model SNPS go to Favorites -> Command Line
- In the command line type select: 123 where 123 is the specific residue (so for a SNP at aa 75, type select: 75)
- Turn the residue into a stick Actions → Atoms/Bonds → Show
- Then, click Tools → Structure Editing → Rotamers
- This will bring up a selection box. Change the rotamer type to the SNP amino acid, click Ok. Click the highest probability rotamer, then under Existing side chain(s) box should be changed to retain to keep the original SNP on there. Click Ok.
- Then highlight the original residue under Actions -> Color
- The changed color (I chose green) is the original, and the branching that remained the same color shows the new residue SNP (hopefully).
How to Check for Clashes/Hindrance
Instructions for mutating a residue, checking for clashes, and structure minimization were derived from NIAID Bioinformatics video tutorials: Part 1 and Part 2
- Steric Analysis was performed on mutated residue to observe clashing residues
- Select mutated residue by holding down “Control” and left clicking on residue. Use up arrow to ensure that entire residue is selected.
- Click Tools -> Surface Binding Analysis -> Find Clashes and Contacts
- On the pop-up window, click "Designate" and check the designated atoms against “all other atoms”. Leave Default Parameters.
- Under “Treatment of Clash/Contact Atoms”, ensure the boxes next to “Select”, “Color”, “Draw pseudobonds of…” and “Write information to reply…” are checked, then click apply.
- Clashing atoms are highlighted in red and psuedobonds between them are highlighted in yellow. Hovering over each residue will show which residue is involved in the clash.
How to Find Energy Minimization
- Hydrogen bonds were removed by Favorites --> Command Line --> and type in "del H"
- Mutated residue and clashes were selected with the up arrow
- Click Tools -> Structure Editing --> Minimize Structure
- Due to time, Steepest Descent Steps was changed to 15, Conjugate Gradient Steps was changed to 0, Fixed Atoms were changed to unselected, and Update Interval was changed to three
- Under the Find Clashes/ Contacts window, Select was deselected, Pseudobonds were changed to red, and Check... after relative motions was selected
- To begin, Minimize was clicked
- OK was pressed to add hydrogens, followed by charges
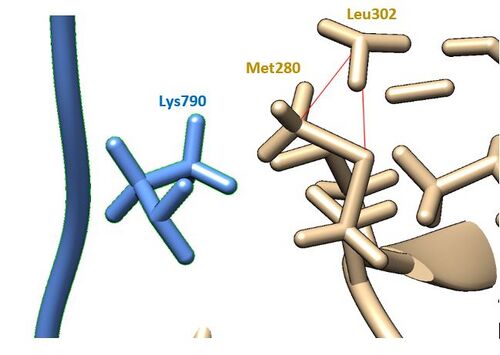
- 2 clashes are shown below, interacting with itself (Leu 302)
Images
V280M
- rs142446494 V280M mutation was visualized using Chimera using the methodology listed above
- this mutation does not appear to cause steric hinderance with neighboring residues, however further steric analysis shows that it will clash with neighboring residues on itself
Default Visualization
- entire residue is highlighted in green, residues in white are original residue (V), residues in yellow are mutated residue (M)
Designated Color Visualization
- blue= TMPRSS2, SARS-CoV-2= magenta, green residue indicates wildtype (V280), blue residue is the mutation (M280)
- Focused in on residues
Clashes/Contacts
- Multiple clashes were shown with neighboring residues of TMPRSS2
- This may prevent proper binding of TMPRSS2 to SARS-CoV-2
- Clashes viewed with hydrogen bonds
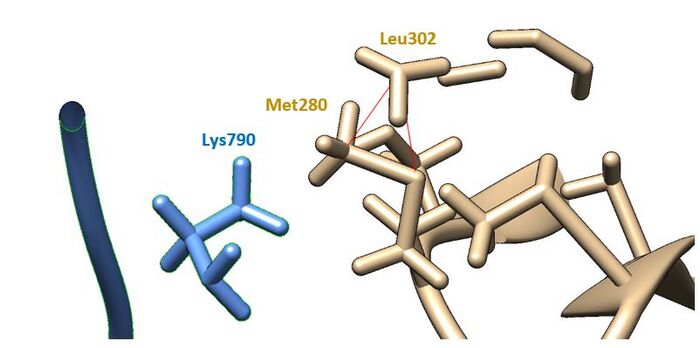
Minimization
- cornflower blue = SARS-CoV-2, tan = TMPRSS2
- 2 clashes can be viewed with the red psuedobonds
ConSurf
Active Sites
Data and Files
Link to SNP Table TMPRSS2 Structure TMPRSS2 and SARS-Cov-2 Interactions TMPRSS2 Gene Map Link to Fall 2020 Research Summary Link to Abstract
Capstone
Annotated Bib
Annika's Annotated Bibliography Jessica's Annotated Bibliography Madeleine's Annotated Bibliography
Outline
Madeleine's Outline Annika's Outline Jessica's Outline
Results
Madeleine's Results (draft) Annika's Results Jessica's Results Draft 1
Discussion
Jessica's Discussion Draft 1 Madeleine's Discussion Draft Annika's Results + Discussion Draft
Introduction
Jessica's Introduction Draft Annika's Introduction Draft Madeleine's Introduction Draft
Materials & Methods
Madeleine's Materials & Methods Draft Annika's Materials & Methods Draft Jessica's Materials & Methods Draft
Appendix
Data & Graphs
- SNP Frequency & Location Table
- Population frequencies of the 18 SNPs of interest were gathered on Excel using ALFA and split according to ALFA defined populations and shown in “All SNPs”. “All SNPs Location” discloses the location and amino acid change of the 18 SNPs, where also recorded from …. “Deleterious SNPs” displays a list of seven of the 18 SNPs that had been classified as “Deleterious” or “possibly damaging” according to SIFT and PolyPhen-2 .
- SNP Interaction Table
- Table displaying the bonding interactions between TMPRSS2 SNPs and SARS-CoV-2 virus. iCn3d was used to visualize the interactions.
- SNP Frequency Graphs
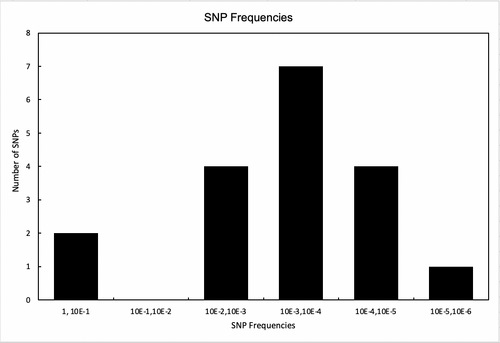
- These Excel sheets group the SNPs based on their frequencies. “No. of SNPs vs. Allele Frequency” plots the SNPs against their frequencies in ranges of 0.0001. “No. SNPs v. 10^x freq.” plots the SNPs in frequencies that correlate with the base 10 of one another. “No. SNPs v. (log(freq))^-1” plots SNPs against the negative inverse of the logarithm (base 10) of the SNP frequencies, grouping them in ranges of 0.1.
- SIFT/PolyPhen2 Results
- This table shows the effect of each of the 18 SNPs amino acid substitution on protein function, according to results from SIFT and PolyPhen--2. SIFT defines SIFT scores range from 0 to 1: the amino acid substitution is considered damaging if the score is less than 0.05 and is tolerated if the score is greater than 0.05. Polyphen-2 scores range from 0 to 1: the amino acid substitution is considered benign if the score is less than 0.05, possibly damaging if the score is between 0.5 and 0.9, and probably damaging if the score is between 0.9 and 1.
Software Files & Images
- Biorender TMPRSS2 Gene Map
- Gene map diagram and Isoform 1 of TMPRSS2. TMPRSS2 is located on Chromosome 21 at q 22.3. It has an enhancer, regulatory region, and promoter located before the transcribed portion of the gene. After splicing, there are 14 exons in Isoform 1. Each of the SNP amino acid substitutions are plotted along Isoform 1, which contains 4 domains.
- Chimera
- Chimera is a software used to visualize molecular structures. It was used to visualize rs142446494 V280M mutation and check for steric hindrance, clashing, and contacts with SARS-CoV-2.
- CLUSPRO Docking Models
- ClusPro was used for protein-protein docking. It was used to dock TMPRSS2 and SARS-Cov-2.
- Haddock Spike Protein TMPRSS2 Summary
- Haddock models the interactions of 2 molecular structures and how they fit together. It was used to model the interaction between TMPRSS2 and the SARS-Cov-2 spike glycoprotein.
- HHpred
- HHpred is a software utilized for detecting remote protein homology and structure prediction, including secondary and tertiary structure. It was used to generate a model and Ramachandran plot of TMPRSS2.
- iCn3D
- iCn3d is a software used to display the 3-D structures of biomolecular structures as well as their interactions and sequences. It was used to visualize the interaction between TMPRSS2 and SARS-CoV-2.
- i-TASSER
- I-TASSER is a protein modeling resource that uses threading to predict protein secondary and 3D models. It was used to generate models for TMPRSS2 and TMPRSS4, and generate a Ramachandran plot.
- Predict Protein Heat Map
- Predict Protein is a software that predicts the effect mutations may have on a protein’s structure and function. It was used to determine the possible effects of the SNPs in the protein.
- RaptorX
- RaptorX is a protein structure prediction server developed that can predict secondary and tertiary protein structures, contacts, solvent accessibility, disordered regions and binding sites. It was used to generate a model and Ramachandran plot of TMPRSS2.
- Swiss Model
- Swiss Model is a protein structure homology modeling software. It was used to generate a model and Ramachandran plot of TMPRSS2.
- TMPRSS2 FASTA files
- FASTA files were found on UnitPro and edited to reflect each SNPs amino acid change. Was input into other softwares to predict protein models and interactions.
Posters, Abstracts, Recordings
- Spring 2021 Undergraduate Research Symposium Poster & Experimental Biology Poster
- TMPRSS2 Frequencies Abstract URS 21
Box Folder
Capstone