IGEM:Hong Kong HKUST/Investigations/ Plasmid Cloning of BFP Generator Using Standard Assembly: Difference between revisions
| Line 77: | Line 77: | ||
===Gel Electrophoresis, Purification, and Nanodrop Test=== | ===Gel Electrophoresis, Purification, and Nanodrop Test=== | ||
Gel Electrophoresis was performed for 30 minutes using 1% agarose gel and midori green as dye. The voltage and current used were 130 V and 400 mA respectively. The expected bands were | Gel Electrophoresis was performed for 30 minutes using 1% agarose gel and midori green as dye. The voltage and current used were 130 V and 400 mA respectively. The expected bands were 2726 bp for J61002-BBa_J23110 and 771 bp for BBa_K592023. After the gel photo was obtained, desired band was taken and purified according to the protocol. The purified band then tested for its concentration and protein and salt contamination by using Nanodrop machine. | ||
===Ligation=== | ===Ligation=== | ||
Revision as of 13:28, 5 June 2016
<html> <body>
Plasmid Cloning of BFP Generator Using Standard Assembly
</body> </html>
Authors
- Winnie Ho
- Claire Moran
- Mike Dorothea
- Sharon Lai
- Olivia Winata
- Luna Eresta Jaya
- Kashish Aggarwal
- Yiu, Stephanie P.T.
Abstract
This training was conducted with the aim to learn the basic skills in producing a desired protein by using DNA recombination techniques such as digestion, ligation and transformation. In this training, constructs were made by inserting a Blue Fluorescent Protein (BFP) generator (BBa_K592023) into a parental plasmid containing a promoter (J61002-BBa_J23110).
The original plasmid containing the promoter (J61002-BBa_J23110) was digested using SpeI-HF and PstI-HF restriction enzymes, while the original plasmid containing the BFP generator (pSB3K3-BBa_K592023) was digested using XbaI-HF and PstI-HF. As a result, five different possible constructs might be produced: the desired construct, a construct with the wrong insert orientation, a construct with two backbones, a construct with two inserts and plasmids that are self-ligated.
Hence, this investigation will explore on methods that can help investigators identify candidate colonies which contain the desired sequence/construct and separate them from the rest. Colony PCR was chosen for colony identification after ligation and transformation. To distinguish the orientation of plasmid cloning, the primers VR and VF2 (which have opposite orientations) were used. Once the PCR products (in the form of amplified, transformed plasmids) were run in gel electrophoresis, the candidates showing the expected band size were concluded to have the desired construct (J61002-BBa_J23110-BBa_K592023). These particular candidates were then inoculated.
Introduction
By using BioBricks Standard Assembly, we can assemble two BioBricks containing our desired parts into the same plasmid, this can then be transformed into a cell and cloned to obtain our desired results. The objective of our project was to create a recombinant cell containing a BFP generator from the BioBricks BBa_J23110 and BBa_K592023. BBa_J23110 is a constitutive promoter, and BBa_K592023 is an intermediate containing a BBa_K592100 Blue Fluorescent Protein and a BBa_B0032 RBS. After combining the two BioBricks by restriction digestion and ligation, then transforming the recombinant plasmid into a competent cell, blue fluorescent protein producing E. coli could be obtained. The details of the BioBricks used are shown in Table 1. BioBricks used in this investigation.
Methods and Materials
BioBricks
Table 1. BioBricks used in this investigation.
| BioBricks | Size (bp) | Plasmid | Description |
| BBa_J23110 | 35 | J61002 | RFP Producing Constitutive Promoter |
| BBa_K592023 | 771 | pSB3K3 | RBS (BBa_B0032) and BFP CDS (BBa_K592100) |
Restriction Enzyme Digestion
The plasmid containing the constitutive promoter (J61002-BBa_J23110) was cut by the restriction enzymes SpeI-HF and PstI-HF and served as the vector in the ligation, while the plasmid containing the BFP generator (pSB3K3-BBa_K592023) was cut by XbaI-HF and PstI-HF and was isolated as an insert. The buffer used was CutSmart and the restriction digestion process was carried out for 1 hour 30 minutes at 37°C. The volumes of the reagents used for this process are described in the following table.
Table 2. Restriction Enzyme Digestion Recipe
| J61002-BBa_J23110 | pSB3K3-BBa_K592023 | |
| DNA Amount (ng) | 300 | 500 |
| Sample DNA Concentration (ng/μl) | 67.14 | 192.3 |
| DNA Sample Volume (μl) | 4.46 | 2.60 |
| Xbal-HF (μl) | - | 0.20 |
| Spel-HF (μl) | 0.20 | - |
| PstI-HF (μl) | 0.20 | 0.20 |
| CutSmart Buffer (μl) | 1.80 | 1.80 |
| ddH20 (μl) | 11.34 | 13.20 |
Gel Electrophoresis, Purification, and Nanodrop Test
Gel Electrophoresis was performed for 30 minutes using 1% agarose gel and midori green as dye. The voltage and current used were 130 V and 400 mA respectively. The expected bands were 2726 bp for J61002-BBa_J23110 and 771 bp for BBa_K592023. After the gel photo was obtained, desired band was taken and purified according to the protocol. The purified band then tested for its concentration and protein and salt contamination by using Nanodrop machine.
Ligation
0.5μL of T4 DNA Ligase was used to ligate J61002-BBa_J23110 with BBa_K592023. The proportion amount of backbone (J61002-BBa_J23110) to amount of insert (BBa_K592023) is 1 to 3.
Transformation
10μLof ligated product was inserted into 50μL of competent cell and put into ice for 20 minutes. After 20 minutes, heat shock and cold shock were performed. After undergo these processes, 1000μL of LB was added into the tube, and the tube was centrifuged for 2 minutes on 6500rpm.
Incubation
Two plates of agar containing the AMP antibiotics were taken out from the fridge, and the transformed bacteria containing the desired plasmid were spreaded on the plates. These plates were incubated overnight.
After colonies were cloned, some of the bacteria were taken as samples for PCR and streaked on new compartmentalized plates that served as back up for another PCR.
Colony PCR
First, mastermix containing 20 folds of 2μL of 10X ThermoPol buffer, 0.5μL of 10mM dNTPs, 0.5μL of VF2 as foward primer and VR as reverse primer, 0.15μL of Taq DNA Polymerase, and 14.35μL of ddH2O. The first step of PCR after the mastermix was added with 2μL template DNA was initial denaturation to burst out the cell for 30 minutes on 95 °C. After that denaturation was done in 30 seconds for 95°C to separate the double stranded DNA. Next step was annealing for 1 minutes and extension for 1 minutes 30 s. The final extension was 3X longer than the extension which becomes 4 minutes 30 s. The final step was holding for around 10 minutes on 10°C.
Gel Electrophoresis
The same step was performed as the previous gel electrophoresis.
Inoculation
5ml of LB containing AMP antibiotics was added to named empty falcon tubes. The colony was inserted to the falcon tubes and incubate overnight.
Miniprep
Miniprep was done according to the protocol. Then nanodrop was performed.
Restriction Check
Restriction check was not applicable in this case because the restriction enzymes we planned to use were denatured, and since the backbone is J61002_J23110, if we use EcorI and PstI the negative results will show similar bands to the experimental setup.
Results
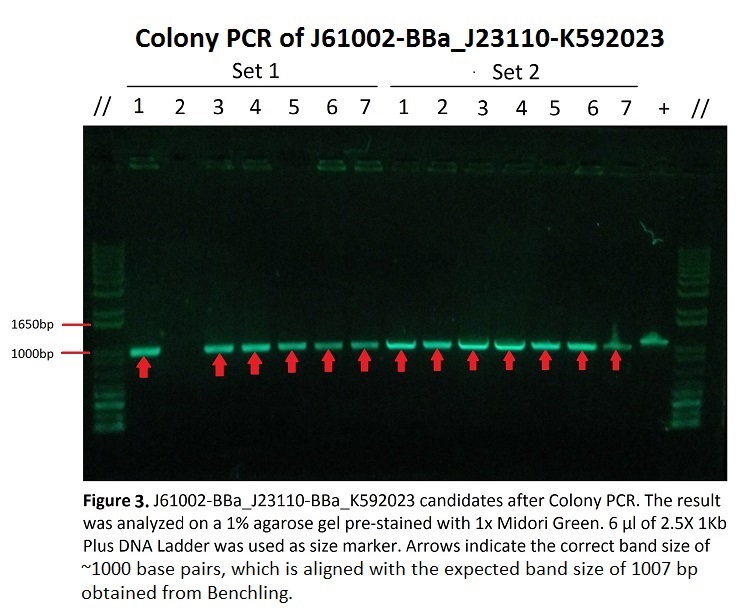
Figure 3 shows Gel Electrophoresis of J61002-BBa_J23110-BBa_K592023 candidates after colony PCR. As seen in the picture, the target bands of ~1000 base pairs are shown, measured by comparison with the DNA ladder (1Kb Plus).
All of the candidate samples from both sets of the experiment showed the expected band, except for Sample 2 in Set 1 which showed no band at all. This, along with the uneven brightness of the bands might have been caused by some random and systematic errors during the experiment, which will be talked about under Discussions.
Discussion
As expected, the plasmid containing BFP generator produced blue colour under UV light. However, there were a couple of failures in the PCR step. On the third time of doing the PCR, bands containing colonies with blue colour appeared on the gel. This indicates that there is a high probability the candidates from the PCR are in fact correct. There are some probable reasons as to why the PCR was not successful. The first reason is there were too many cells in the samples so the gel could not run properly. The second reason is human error when mixing the master mix. The third one is that not enough Taq polymerase was added.
Improvements
The process of making the master mix must be done carefully to reduce the chance of failure, and sufficient Taq polymerase should be added overtime when doing the PCR. Proper restriction check and sequencing should be done to further verify the candidates.
Conclusion
In conclusion, the BFP producing E. coli was successfully created. The compositions of each process must be precise and exact so the desired result can be obtained.