BISC209/F13: Lab2: Difference between revisions
Tucker Crum (talk | contribs) |
Tucker Crum (talk | contribs) |
||
| Line 76: | Line 76: | ||
Adapted from Schallenberg, M., Jacob, F. and Joseph B. R. (1989) Solutions to Problems in Enumerating Sediment Bacteria by direct counts. Applied & Environmental Microbiology. p. 1214-1219.<BR><BR> | Adapted from Schallenberg, M., Jacob, F. and Joseph B. R. (1989) Solutions to Problems in Enumerating Sediment Bacteria by direct counts. Applied & Environmental Microbiology. p. 1214-1219.<BR><BR> | ||
=='''RICHNESS OF A MICROBIAL SOIL COMMUNITY: METABOLIC DIVERSITY: <BR> | =='''RICHNESS OF A MICROBIAL SOIL COMMUNITY: METABOLIC DIVERSITY: <BR>Structural Diversity in Cultured Bacteria from a Soil Community''== | ||
<font size="+1">Using General Purpose | <font size="+1">Using General Purpose Media for Isolation of Soil Bacteria in a Mixed Population</font size="+1"><BR> | ||
Please watch the YouTube video on streaking for isolation and pay attention to your instructor's demonstration: http://www.youtube.com/watch?v=eyoW18Fzb3o<BR> | Please watch the YouTube video on streaking for isolation and pay attention to your instructor's demonstration: http://www.youtube.com/watch?v=eyoW18Fzb3o<BR> | ||
Directions for [[BISC209/F13: Streaking for Isolation| Streaking for Isolation]] are found in the [[BISC209/F13:Protocols| Protocols]] section of this wiki. <BR><BR> | Directions for [[BISC209/F13: Streaking for Isolation| Streaking for Isolation]] are found in the [[BISC209/F13:Protocols| Protocols]] section of this wiki. <BR><BR> | ||
'''Streaking for Isolation from a | '''Streaking for Isolation from a Soil Extract:'''<BR> | ||
Each '''team''' of students will '''make a soil extract''' from one gram of their | Each '''team''' of students will '''make a soil extract''' from one gram of their soil sample. | ||
<li>After recording the weight of the individual aluminum weigh boats, combine the dry soil into one weighing boat, mix gently, and then weigh out 1 gram of the mixed oven dried soil on waxed weighing paper. | <li>After recording the weight of the individual aluminum weigh boats, combine the dry soil into one weighing boat, mix gently, and then weigh out 1 gram of the mixed oven dried soil on waxed weighing paper. | ||
<li>Add the 1g of mixed dried soil to 100ml of sterile water. This is the equivalent to a 1/100 dilution. | <li>Add the 1g of mixed dried soil to 100ml of sterile water. This is the equivalent to a 1/100 dilution. | ||
| Line 113: | Line 113: | ||
<li>We will select colonies of other potential spore-forming bacteria in lab 3. </ul> | <li>We will select colonies of other potential spore-forming bacteria in lab 3. </ul> | ||
<BR> | <BR> | ||
'''Isolation of | '''Isolation of Bacteria by Spread Technique:'''<BR> | ||
<ul> | <ul> | ||
<Li>Transfer 100 µL of soil extract onto a pre-labeled plate of | <Li>Transfer 100 µL of soil extract onto a pre-labeled plate of medium. | ||
<LI>Using a sterile disposable spreader cover the surface of the plate with the inoculum. | <LI>Using a sterile disposable spreader cover the surface of the plate with the inoculum. | ||
<LI>Wait one minute to allow the inoculum to absorb into the agar. | <LI>Wait one minute to allow the inoculum to absorb into the agar. | ||
<li>Invert, and incubate the plate at RT. | <li>Invert, and incubate the plate at RT. | ||
<li>Check your plate for colonies daily and record your descriptions of the texture and shape of the colonies that appear. If any colonies arise that look tough and leathery or appear as "little, powdered-sugar volcanos", use the tip of a sterile toothpick to pick up a small but visible amount of growth (being careful not to touch anything but the tip of the colony) and "spread" the bacterial growth onto section 1 of a new | <li>Check your plate for colonies daily and record your descriptions of the texture and shape of the colonies that appear. If any colonies arise that look tough and leathery or appear as "little, powdered-sugar volcanos", use the tip of a sterile toothpick to pick up a small but visible amount of growth (being careful not to touch anything but the tip of the colony) and "spread" the bacterial growth onto section 1 of a new plate. (Inoculate one colony/plate.) Use your loop to isolation streak sections 2-4. Don't forget to flame your loop between sections! | ||
<li>We will select colonies | <li>We will select colonies again in lab 3. </ul><BR><BR> | ||
Over the next few weeks you will continue to sub-culture onto new plates, using your best isolation streak technique. Your goal is to continue to streak out ONE CFU until you are sure that all the bacterial growth in a colony comes from a single mother cell (pure culture). In subsequent labs you will make a bacterial smear and do a Gram stain of these genetically identical bacteria and you will perform other tests from freshly pure cultures to explore the physical and metabolic characteristics of this isolate. <BR><BR> | Over the next few weeks you will continue to sub-culture onto new plates, using your best isolation streak technique. Your goal is to continue to streak out ONE CFU until you are sure that all the bacterial growth in a colony comes from a single mother cell (pure culture). In subsequent labs you will make a bacterial smear and do a Gram stain of these genetically identical bacteria and you will perform other tests from freshly pure cultures to explore the physical and metabolic characteristics of this isolate. <BR><BR> | ||
Some bacteria often form tough leathery colonies, so transfer of these bacteria to new media to start a sub-culture is sometimes difficult. The powdery area may be spores which would be interesting to visualize later. To isolate them try to "break off" a piece of the colony with your sterile loop or with a sterile toothpick and transfer that whole piece of a colony onto zone one of the new plate. Then use your loop for streaking out the other zones. The tiny spores on the surface of the colony are likely to transfer to the next plate or tube when you work with it. (That's a good thing this time.) <BR><BR> | |||
Your team will obtain additional Streaking for Isolation practice by following the directions for | Your team will obtain additional Streaking for Isolation practice by following the directions for ISOLATION using the spread plates that you inoculated with your soil extract last week. This process will continue your attempt to find a diverse group of interesting bacteria in your soil community. <BR><BR> | ||
The directions for [[BISC209/F13: Aseptic Transfer| Aseptic transfer: Broth to Broth and Broth to Plate]] are found in the [[BISC209/F13:Protocols| Protocols]] section of the wiki. <BR> | The directions for [[BISC209/F13: Aseptic Transfer| Aseptic transfer: Broth to Broth and Broth to Plate]] are found in the [[BISC209/F13:Protocols| Protocols]] section of the wiki. <BR> | ||
==CLEAN UP== | ==CLEAN UP== | ||
Revision as of 21:56, 20 August 2013
LAB 2: Enumerating Microbial Members & Isolating Bacterial Members of a Soil Community
Today you will complete your first investigative goal in our semester long project: assessing abundance of a soil microbial community by enumerating its microbes by two methods, one culture dependent and one culture independent. Today you will also continue acquiring and practicing skills used by working microbiologists. You will complete your tests for abundance today. The results of the fluorescent DNA stain are available and your plate counts are ready to assess. In addition, you will continue to isolate a few bacteria of interest from your soil community in order to show evidence for richness (diversity) and potential for co-operative and competitive behaviors among community members. Next week we will begin testing the whole soil microbial community : a culture-dependent assessment of carbon source utilization diversity.
Background to Abundance Testing: The Great Plate Count Anomaly
Advances in molecular tools for gene sequencing and in other types of microorganism identification have dramatically expanded our knowledge of the contribution of microbes in their (and our) environment. It is estimated that 99.9% of microbes are "unculturable" - that is, currently not able to be cultured by traditional methods! The large number of these uncultured bacteria are responsible for the, so-called, "Great Plate Count Anomaly": the recognition that many more bacteria are present than appear as colonies on agar plates. Culture-independent estimates of the number of bacteria in a gram of soil are 109 - this is several hundred to 9,000 orders of magnitude greater than the number derived from culture-dependent methods. It has been speculated that there might be 10 billion species of bacteria on Earth!
Abundance Test 1: Finishing the Standard Plate Count
Last week you started a standard plate count of the culturable microbes in your soil sample on dilute nutrient agar. Today you will complete that plate count to get one kind of enumeration of the microorganisms in your soil community. Find the plate(s) among those of your serial dilution that contains 30-300 colonies. You will not assess plates with well over 300 colonies or under 30; they should be designated as "invalid" in your lab notebook and on the bottom of the plate. Count all the surface and subsurface colonies on the replicate valid plates (those with 30-300 colonies). The colonies can be more easily counted by using a Quebec Colony Counter which allows proper illumination, a grid overlay, and slight magnification of the plate surface. (There are two colony counters in the lab.)
Calculating the number of colony forming units (CFU) per gram of soil
For each valid plate counted, divide the total number of colonies counted by the amount of inoculum plated times the dilution factor of that plate to obtain the number of bacteria per gram of soil.
number CFU/(dilution plated*dilution factor) = number of CFU/gram
For example, if you counted 150 colonies on the 10-6 plate the calculation is:
150/(0.1ml inoculum*1X10-6dilution)= 150X107 which in scientific notation is written as 1.5X109 CFU/gram
Record the number of CFU/ gram of wet soil in your lab notebook and enter your data on the course spreadsheet on the instructor's computer at the front of the lab. Calculate the average and standard deviation for the plates from your sample site. Add your average and standard deviation data to the chalkboard for comparison with the other soil sampling sites. Note that this number is the average colony forming units/ gram WET soil for your sample site.
Soil bacteria are usually not recorded as number of colony forming units (CFU) in 1 gram of WET soil but instead as per gram of DRY weight. Therefore, you will need to figure out your counts as DRY weight. Please weigh each of the three 1 gram samples that you left last week for oven drying. Average the new dry weights. The weights should be considerably less than 1 gram. Save the 3 dried soil samples after you weigh them, you will need them again today.
Determine the % change in soil weight by subtracting the average dry weight from 1 g wet weight divided by the wet weight, then times 100.
[(wet weight - dry weight)/wet weight] * 100 = % change
Use this % change to convert the CFU per gram wet soil to CFU per gram dry soil. For example: If your dry weight soil averages 0.75 grams, then (1g-0.75g)/1g * (100) = 25% change. The number of microbes should be 25% higher than the number calculated above/gram of wet weight. Calculate 25% (or whatever your conversion factor is) of your CFUs and add that number to the CFUs/ gram of wet weight soil.
Record the number of CFU/ gram DRY weight soil in your lab notebook and add this information to the chalk board. If you are unsure of the accuracy of your calculations of CFUs per gram of wet soil weight and per gram of dry weight, check with your instructor. You will need accurate counts for some of our future testing.
Things to Consider:
Why did we perform so many dilutions when we set up our plate count in Lab 1?
Why should you have only one dilution with 30-300 colonies?
Why did we make up to 4 replicates per site?
Do you expect the results of this enumeration of soil microbes in your community to be the same, lower, or higher than the results of our other enumeration test (the DAPI DNA stain)? Why?
Seal the edges with parafilm of the valid dilute nutrient agar plate and store the plate in the cold room at the end of lab. Don't discard any other plates yet.
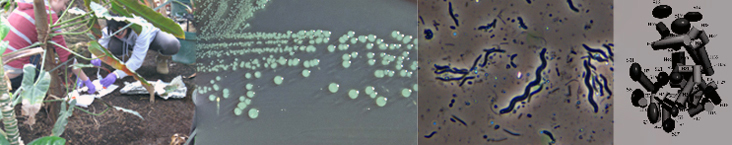
Abundance Test 2: Enumeration of Community Soil Microorganisms by Direct Count of Microbial DNA Stained & Viewed by Fluorescence Microscopy
You can directly count a random sample of microbes from the soil extract that you prepared last week and then extrapolate the number per gram of soil. To make the microbes easier to count, your lab instructors stained the nucleic acids of your soil community microbes (not just the bacteria) with a fluorescent 4'-6-Diamidino-2-phenylindole (DAPI) DNA stain. All the microbes in a 1ml aliquot of the 1% soil extract that you prepared last week were transferred in a Poisson distribution to a small piece of filter paper. She photographed the discreet bright "spots" of fluorescent DNA in several different areas of the filter paper distribution using fluorescence microscropy. These photomicrographs will be made available to you and your partners today. You will each count the discreet "spots" as individual microorganisms (one fluorescent genome/cell) from one area and perform the calculations described below to assess the total microbial concentration in this culture-independent enumeration of your soil community's microorganisms. Compare this number to the calculation of CFUs/gram of wet soil (a culture-dependent assessment) that you will also obtain today. Compare the two counts that, in theory, should be the same since we are using two methods to figure out the same thing: how many microbes per gram comprise your soil community. The answer will provide evidence for one of our investigative goals: abundance or microorganisms in your soil community. What does it mean if your two experimental methods don't give the same answer?
DIRECT COUNT OF MICROBIAL CELLS USING DAPI DNA: PROTOCOL
NOTE: The staining and imaging were performed by your lab instructor last week according to the protocol described below (provided here for reference only). YOU WILL NOT PERFORM THIS WORK IN LAB TODAY!
DAPI aqueous solution Stock conc. is 1mg/ml : 4'-6-Diamidino-2-phenylindole (DAPI) is known to form fluorescent complexes with natural double-stranded DNA.
- If the samples will not be processed for a while: You need a final concentration of 4% parformaldahyde solution that will act as a preservative for the microbial cells in your soil extract samples. Ask your instructor about this step.
- Add 5μL of 1mg/ml DAPI stock (Thermo scientific prod. # 62248) stain to 1mL of a fresh 1% diluted soil extract so that the effective concentration of DAPI is 5 μg/mL (1μg/mL may work as well).
- Incubate at 4°C for 20 min in the dark.
- Set up a vacuum flask and filter apparatus (125 ml side-arm flask, Borosilicate base and fritted glass filter support with rubber stopper).
- Carefully place a 0.2μM glass fiber filter (Isopore membrane GTTP02500) onto the fritted glass filter support.
- Add the borosilicate glass funnel onto the base and clamp the two sections together using an anodized aluminum spring clamp.
- Rinse the filter once with 1 mL sterile deionized water using a vacuum pump to provide 178mm Hg of force. Wait until all the water is removed.
- Turn off the vacuum, gently break the vacuum by loosening the rubber stopper, re-tighten, and carefully transfer the 1mL of DAPI stained extract (made in step 1) onto the 0.2μM glass fiber filter.
- Turn the vacuum on and wait until all the solution is filtered.
- Rinse with 1 ml sterile deionized water to remove any background DAPI.
- Dissassemble the funnel, gently breaking the vacuum by loosening the rubber stopper.
- Use forceps to carefully remove the filter and place it on a microscope slide.
- Add 1 drop of Fluoro-Gel with Tris buffer (Electron Microscopy Sciences Cat #17985)and a coverslip.
- Use the Nikon Eclipse 80i fluorescent microcope in L318C to view and photograph the DNA at 1000x magnification.
- Use the Nikon NIS-Elements Imaging software, to photograph a field of view in white light (this allows you to see relative size and shape of the organisms in the sample, remember some will be eukaryotes), save this image, then do the same using the UV light at 350nm excitation wavelength (filter #1) so you can visualize the fluorescently labled DNA.
- Save all images to a 209-2012 file folder.
- Count the unique spots of blue fluorescence; each indicates a soil microbe's genetic material (chromosome or nucleus). It is assumed that the bacteria are arranged in a Poisson distribution. For the most accurate counts, 20 fields or 400 bacteria should be enumerated to determine the number of bacteria per ml.
COUNTING & CALCULATIONS:
The area of each field of view at 1000X using the Fluorescent scope is 10487 μmeters2. The area is determined for the microscope used, in our case, a NIKON 80i fluorescent microscope. The diameter of the filterable section of the borosilicate apparatus is 17 mm (8500 μmeter radius). Therefore, the area is 2.28 X 108μmeter2. Multiply the number of microorganisms counted on the photomicrograph by a factor of 2.17X104 (2.28 X 108μm2 divided by 1.0487 x 104μmeters2) to determine the number of organisms found in 1mL of filtrate of extract. Then correct for the 1:100 dilution factor of filtrate which is the number of organisms in 1 gram of wet soil. Convert this to the number of organisms in the community in 1 gram of dry weight soil.
Post your mean (average of the areas of the filter counted) number of microbes/gm of soil to the spread sheet on the instructor's computer and on the board (all calculations must also be in your lab notebook!) from both the culture-dependent and culture-independent enumerations. Report them as the estimated number of microorganisms in 1gm of wet weight soil and 1gm of dry weight soil. Record these numbers in your lab notebook in scientific notation and as numbers (with an amazing number of zeros). Consider the relative insignificance of one gram of anything and the enormity of the number of microbes that thrive in a gram of soil! How is it possible that each of the microbial members of such a soil community can find a niche, obtain all the nutrients needed to grow and reproduce, and contribute to overall health of the soil and to the community of microorganisms?
Now that we have some sense of the abundance of microbes in a soil community we can move on to investigating the richness (diversity) and the co-operative and competitive behaviors that maintain it.
Adapted from Schallenberg, M., Jacob, F. and Joseph B. R. (1989) Solutions to Problems in Enumerating Sediment Bacteria by direct counts. Applied & Environmental Microbiology. p. 1214-1219.
'RICHNESS OF A MICROBIAL SOIL COMMUNITY: METABOLIC DIVERSITY:
Structural Diversity in Cultured Bacteria from a Soil Community
Using General Purpose Media for Isolation of Soil Bacteria in a Mixed Population
Please watch the YouTube video on streaking for isolation and pay attention to your instructor's demonstration: http://www.youtube.com/watch?v=eyoW18Fzb3o
Directions for Streaking for Isolation are found in the Protocols section of this wiki.
Streaking for Isolation from a Soil Extract:
Each team of students will make a soil extract from one gram of their soil sample.
Aseptic transfer and streaking for isolation are crucially important skills in microbiology. When you are ready to streak out your first plate (after you have read the directions carefully, asked questions to clarify any confusion, and practiced your best technique on an empty plate or piece of paper), call your instructor over and ask her to watch you streak out your first plate. She will give you pointers to enhance your success at repeating this technique on a second plate. Your goal is to have well-separated single colonies when you evaluate your streaked plates next week. Label the plates with your initials, date, lab section, soil sample identifier information, and medium name. Incubate one plate at room temp and one plate at 30°C in the racks assigned to your group or Lab section.
Enrichment & Isolation of endospore-forming bacteria from oven dried soil extract on Glycerol Yeast Extract (GYEA) Medium: 0.5% (v/v) Glycerol, 0.2% Yeast Extract, 0.1%K2HPO4(Dipotassium phosphate), 1.5% Agar
Endospore forming genera include Bacillus and Streptomyces as well as other members of the Actinomycetes family. Many of these bacteria are antibiotic producers.
Oven drying will have killed most of the vegetative non-spore forming microbes. GYEA medium is particularly selective for the Streptomyces group of endospore formers because of the osmotic pressure obtained from the high concentration of glycerol. Few bacteria can thrive in GYEA which favors the growth of members of the Streptomyces group of Gram positive bacteria.
In pairs set up enrichment/selection cultures for spore-forming bacteria on two plates of glycerol yeast extract agar (GYEA) following the directions below (also found in the Enrichment Media for the Isolation of Soil Bacteria in a Mixed Population: Finding Spore Forming Bacteria in the Protocols: Culture Media section of this wiki).
Isolation of Spore-formers by isolation streak:
- You will use a new sterile cotton swab dipped into the oven-dried-soil extract to Swab section 1 of a pre-labeled plate of glycerol yeast medium (GYEA).
- Wait one minute to allow the inoculum in section 1 to absorb into the agar before you switch to your inoculating loop to streak from section 1 into section 2, following the steps for Streaking for Isolation in the Protocols section of this wiki.
- Flame your loop and isolation streak section 2.
- Flame and Repeat to streak sections 3 and 4.
- Invert, and incubate the plate at RT.
- Check your plate for colonies daily and record your descriptions of the texture and shape of the colonies that appear. If any colonies arise that look tough and leathery or appear as "little, powdered-sugar volcanos", use the tip of a sterile toothpick to pick up a small but visible amount of growth (being careful not to touch anything but the tip of the colony) and "spread" the bacterial growth onto section 1 of a new glycerol yeast plate. (Inoculate one colony/plate.) Use your loop to isolation streak sections 2-4. Don't forget to flame your loop between sections!
- We will select colonies of other potential spore-forming bacteria in lab 3.
Isolation of Bacteria by Spread Technique:
- Transfer 100 µL of soil extract onto a pre-labeled plate of medium.
- Using a sterile disposable spreader cover the surface of the plate with the inoculum.
- Wait one minute to allow the inoculum to absorb into the agar.
- Invert, and incubate the plate at RT.
- Check your plate for colonies daily and record your descriptions of the texture and shape of the colonies that appear. If any colonies arise that look tough and leathery or appear as "little, powdered-sugar volcanos", use the tip of a sterile toothpick to pick up a small but visible amount of growth (being careful not to touch anything but the tip of the colony) and "spread" the bacterial growth onto section 1 of a new plate. (Inoculate one colony/plate.) Use your loop to isolation streak sections 2-4. Don't forget to flame your loop between sections!
- We will select colonies again in lab 3.
Over the next few weeks you will continue to sub-culture onto new plates, using your best isolation streak technique. Your goal is to continue to streak out ONE CFU until you are sure that all the bacterial growth in a colony comes from a single mother cell (pure culture). In subsequent labs you will make a bacterial smear and do a Gram stain of these genetically identical bacteria and you will perform other tests from freshly pure cultures to explore the physical and metabolic characteristics of this isolate.
Some bacteria often form tough leathery colonies, so transfer of these bacteria to new media to start a sub-culture is sometimes difficult. The powdery area may be spores which would be interesting to visualize later. To isolate them try to "break off" a piece of the colony with your sterile loop or with a sterile toothpick and transfer that whole piece of a colony onto zone one of the new plate. Then use your loop for streaking out the other zones. The tiny spores on the surface of the colony are likely to transfer to the next plate or tube when you work with it. (That's a good thing this time.)
Your team will obtain additional Streaking for Isolation practice by following the directions for ISOLATION using the spread plates that you inoculated with your soil extract last week. This process will continue your attempt to find a diverse group of interesting bacteria in your soil community.
The directions for Aseptic transfer: Broth to Broth and Broth to Plate are found in the Protocols section of the wiki.
CLEAN UP
1. All culture plates that you are finished with should be discarded in the big orange autoclave bag near the sink next to the instructor table. Ask your instructor whether or not to save stock cultures and plates with organisms that are provided.
2. Culture plates, stocks, etc. that you are not finished with should be labeled on a piece of your your team color tape. Place the labeled cultures in your lab section's designated lab area at room temperature (RT), the walk-in 30°C room, or the walk in cold room. Place any tubes in a labeled rack. If you have a stack of plates, wrap a piece of your team color tape around the whole stack and label it with your initials and the date.
3. Remove tape from all liquid cultures in glass tubes. Then place the glass tubes with caps in racks by the sink near the instructor's table. Do not discard the contents of the tubes.
4. Glass slides or disposable glass tubes can be discarded in the glass disposal box.
5. Make sure all contaminated, plastic, disposable, serologic pipets and used contaminated micropipet tips are in the small orange autoclave bag sitting in the plastic container on your bench.
6. If you used the microscope, clean the lenses of the microscope with lens paper, being very careful NOT to get oil residue on any of the objectives other than the oil immersion 100x objective. Move the lowest power objective into the locked viewing position, turn off the light source, wind the power cord, and cover the microscope with its dust cover before replacing the microscope in the cabinet.
7. If you used it, rinse your staining tray and leave it upside down on paper towels next to your sink.
8. Turn off the gas and remove the tube from the nozzle. Place your bunsen burner and tube in your large drawer.
9. Place all your equipment (loop, striker, sharpie, etc) including your microfuge rack, your micropipets and your micropipet tips in your small or large drawer.
10. Move your notebook and lab manual so that you can disinfect your bench thoroughly.
11. Take off your lab coat and store it in the blue cabinet with your microscope.
12. Wash your hands.
13. See you next time!
Assignment
Graded Assignment:
The directions for this assignment found at: Lab 2 Assignment: Assignment 2.
Do before next lab:
1. Someone from your team must go to your sample site and collect a new soil sample as you did in Lab 1. On the day of Lab 3, BEFORE lab begins, stop by the lab to pick up a plastic bag containing materials for collecting a new soil sample from your sampling site. This time your group will only need enough soil to fill half of a sterile, 15ml, orange capped, conical tube. Make sure you get the sample for THE SAME sampling site as your original sample. Again avoid the top few millimeters of surface soil and wear gloves to avoid contaminating the sample with bacteria from your skin. You probably won't need to use the corer; a disinfected metal spatula or even a spoon should work fine. Bring your new soil sample to lab with you and please be on time.
2. Goal for Cultured Bacteria Isolation: Characterize a variety of spore forming, nitrogen cycling, or other interesting bacteria in a soil community
One goal in this project is for your group to end up with pure cultures of 16 unique soil bacteria (4 per person) of the groups we enriched and selected for (antibiotic producers, endospore formers, contributors to the nitrogen cycle) and from other random groups. Work with your partners over the next several weeks so that you each choose different looking bacterial colonies to isolate to pure culture. Make sure that some come from all the different media. Your goal is for each team member's subset to be unique and for the whole group sample to be as diverse as possible.
Note that isolation of desired bacteria from mixed culture is challenging. Your best technical and organizational skills are required. You will be expected to come in outside of lab time, on your own, to start, continue or complete the process of isolating to pure culture. We will make every attempt to make the media and reagents that you require available when you need them, but that availability requires advance planning on your part as well as on ours. Communicating your needs or desires well in advance of time of use, will reduce frustration and speed up the process. Remember that because this is an investigative, project based lab course, your instructors do not know the identity of the bacteria you are culturing from your soil habitat. The success of this project is in your hands.
Links to Labs
Lab 1
Lab 2
Lab 3
Lab 4
Lab 5
Lab 6
Lab 7
Lab 8
Lab 9
Lab 10