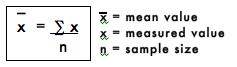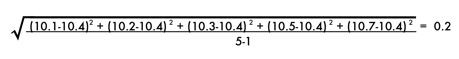BISC110/F13: Series 1 Lab 3 Tetrahymena Investigation: Difference between revisions
| Line 129: | Line 129: | ||
6.0µm Yellow <BR> | 6.0µm Yellow <BR> | ||
Polybeads are packaged as 2.5% aqueous suspension (water) and will be diluted to a working concentration of 0.08% prior to lab. This concentration approximately equals the working concentration of carbon particles in 1% India ink. <BR><BR> | Polybeads are packaged as 2.5% aqueous suspension (water) and will be diluted to a working concentration of 0.08% prior to lab. This concentration approximately equals the working concentration of carbon particles in 1% India ink. <BR><BR> | ||
== Laboratory Cleanup == | == Laboratory Cleanup == | ||
Revision as of 17:41, 11 June 2013
Objectives
In this lab you will learn:
- Scientific observation
- Measurement of specimens using the micrometer
- Experimental design
- Effective Figure design
Series 1 Lab 3 Tetrahymena pyriformis Phagocytosis
Adapted from Bozzone, M.D., and D.A. Martin 2000. An experimenal system to study phagocytosis. Pages 405-415, in Tested studies for laboratory teaching, Volume 21 (S.J. Karcher, Editor). Proceedings of the 21st Workshop/Conference of the Association for Biology Laboratory Education (ABLE).
In this laboratory, you will study Tetrahymena phagocytosis. Phagocytosis is the process by which cells ingest large particles. Unicellular organisms such as Tetrahymena pyriformis carry out phagocytosis in order to obtain nutrients. During the process of phagocytosis, particles are internalized in an endocytic vesicle called a phagosome. The phagosome ultimately fuses with the lysosome so that nutrients can be broken down and used by the cell. In many multicellular organisms, only specialized cells are capable of phagocytosis. Instead of carrying out phagocytosis to obtain nutrients, human phagocytic cells called macrophages are capable of recognizing and engulfing pathogens or abnormal cells. In this lab, you will use Tetrahymena pyriformis as a model organism for the study of phagocytosis. In order to visualize phagocytosis in Tetrahymena, we will provide the cells with India ink as a “food” source. In addition to making scientific observations in this lab, you will also have the opportunity to learn some of the ways that scientific data are evaluated and presented.
PART I: Concepts in Experimentation
A. Accuracy and Precision
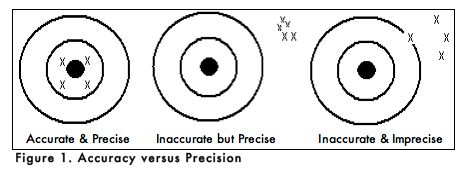
When carrying out an experiment, it is important to do the experiment more than one time to be sure that your measurements are accurate and as precise as possible. Accuracy is the degree of agreement between the measured value of a particular component and the accepted true value. Precision is defined as the degree of agreement between replicate measurements of the same component. Precise measurements do not necessarily indicate accurate measurements. Figure 1 uses targets to illustrate the difference between accuracy and precision. If the bull’s eyes in the targets below represent the true value, then the darts that are the closest to the center of the target are the most accurate. Darts that are not near the center of the target are not accurate even though they may be very closely grouped together indicating good precision. Finally, the darts that are not near the center or grouped together are neither accurate nor precise. Measured values that are both accurate and precise are the goal of all analytical procedures.
B. Statistical Parameters
For various reasons, replicate analytical measurements do not always result in exactly identical values, and so statistical calculations are often used to evaluate analytical data. The validity of these calculations is linked to the number of replicate measurements. Generally, a high number of measured values will yield the most valid statistical evaluation. A short explanation of several elementary statistical parameters on replicate measurements are provided below.
1. Median Value
If all the replicate values for a series of measurements were ordered from the lowest value to the highest value, then the midpoint value is the median. For example, if a series of measured millimolar (mM) concentrations of NaCl are 10.1, 10.2, 10.3, 10.5, and 10.7mM, the median value would be 10.3mM.
2. Range of Values
The range is the difference between the highest measured value and the lowest measured value in a series of replicate measurements and is a simple indicator of precision. A wide range in replicate measured values indicates poor precision. The range is calculated by subtracting the lowest replicate value from the highest. For the above concentrations of NaCl, the range would be calculated as:
3. Mean Value
The mean value is simply the arithmetic average of all the replicate values and is generally considered to be more accurate than any individual measured value. The mean is calculated by adding all the measured values and dividing by the number of measured values or sample size. The mean value is represented mathematically as:
The mean of the NaCl concentrations from step #1 is calculated as follows:
4. Standard Deviation
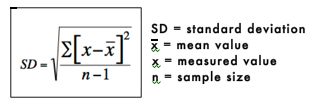
The standard deviation is a measure of how spread out or variable the values are. The standard deviation is calculated by subtracting the mean from each value and then squaring and summing these values. The resulting value is divided by n-1, where n is the number of values. The square root of these values is the standard deviation. The standard deviation is represented mathematically as:
The standard deviation of the sodium chloride values is calculated as follows:
5. Practice Data Analysis
Imagine that you exposed Tetrahymena to ink and observed the number of ink filled vacuoles in 15 cells at both 10 minutes and 30 minutes. You obtained the following data for the number of filled vacuoles at each time point:
Calculate the means and standard deviations of ink filled vacuoles at 10 and 30 minutes. Refer to the Generating Graphs and Error Bars with Microsoft Excel instructions found below or in Appendix A and plot the data in a bar graph with time on the X axis and the mean number of filled vacuoles on the Y axis. Use error bars to denote the standard deviation.
Generating Graphs and Error Bars with Microsoft Excel
- To launch Excel, click on the icon in the dock at the bottom of the screen. An Excel Workbook will open. If Microsoft Excel is already open, select New Workbook from the File menu to display a new spreadsheet.
- To determine the mean and standard deviation, write Average in cell A2 and Standard Deviation in cell A3. List variable 1 in cell B1, variable 2 in cell C1… Now enter the number of filled vacuoles per cell for variable 1 in cells B4, B5, B6... Enter the number of filled vacuoles per cell for variable 2 in cells C4, C5, C6…
- Be sure that the formula builder bar under view is selected.
- To determine the average value for variable 1 (10 min), click on cell B2, press the = symbol, click on the upper left pull down menu and select AVERAGE, highlight cells B4-B18 and press return. The average value should now appear in cell B2.
- To determine the standard deviation for variable 1, click on cell B3, press the = symbol, click on the upper left pull down menu and select STDEV, highlight cells B4-B18 and press return. The standard deviation value should now appear in cell B3.
- To determine average and standard deviation values for variable 2 (30 min), highlight cells B2 and B3, select the lower right square on the highlighted box with the black + symbol, drag to the right until you reach cells C2 and C3, and then release the mouse. The average and standard deviation values for variable 2 should now appear in cells C2 and C3.
- To make your graph, click on cell A1. While keeping the mouse clicked, drag mouse to cell C3 so that cells A1, A2, B1, B2, C1, and C2 are all highlighted.
- Click on the Charts tab below the toolbar. A gallery of chart types will appear below. Press the Column button below to display the appropriate charts in this so-called Elements Gallery.
- To select the proper chart type from the Elements Gallery, press the “Clustered Column” thumbnail (all the way to the left). A chart will appear on the worksheet.
- Open the Formatting Palette by pressing on the Toolbox icon. (If this doesn't work, try clicking on Format and then Data Series.) Under Chart Options you can label the axes and provide a title.
- To add error bars, double click on the columns on your graph. The format data series box will appear. Select Error Bars on the left. Under display, select Both. Under error amount, select Custom and Specify Value. The Custom Error Bars chart will appear. Clear any values in the Positive Error Value box and then highlight the standard deviation values that you calculated previously. Do the same with the Negative Error Values. Click OK twice.
- The chart may be created as an object in the worksheet, or it may be displayed as a new sheet. Under the Chart menu select Move Chart... to choose between these two options.
- Double-clicking on either axis will allow you to change the scale, font and other features of the axis.
PART II: Ink and Tetrahymena Behavior
You will be provided with 1% India ink and live Tetrahymena for this lab. You will add 1% ink to the Tetrahymena and make observations. Based on your careful observations, you will collect some data to quantify Tetrahymena phagocytosis in the presence of India ink.
A. Observations of Tetrahymena pyriformis Exposed to India Ink
- In a microcentrifuge tube, add 50 μL of 1% ink solution and then add 50 μL of live Tetrahymena. Mix gently and record the time.
- What is the effective concentration of the ink after the addition of Tetrahymena?
- Add 20 μL of the Tetrahymena pyriformis and ink solution that you made in step 1 to a glass slide. Add a cover slip and view using the microscope. If you need to review how to focus the microscrope, refer to the directions in Lab 2. You may need to adjust the field diaphragm or to lower the condenser a bit to see the cellular structures clearly. Record your observations in your notebook.
- Do you see any ink particles inside the Tetrahymena? Be sure to note the time as soon as you notice an ink filled phagocytic vacuole forming. Continue observing the Tetrahymena. Watch the Tetrahymena for 15 minutes, noting how many vacuoles form and where those vacuoles are in the cell. Do they move? Are the vacuoles uniform in size?
B. Time course experiment of Tetrahymena pyriformis Exposed to India Ink
After you have had a chance to observe the "Tetrahymena" in the presence of ink, you will do an experiment in which you collect data to present in the form of two figures.
- Put on a pair of gloves.
- To begin your experiment, add 50 μL of 1% ink solution and then add 50 μL of live Tetrahymena. Mix gently and record the time.
- After 0, 5, and 15 minutes, add 20 μL of the Tetrahymena and ink solution to a clean labeled microcentrifuge tube. IN THE HOOD, add 10 μL of 3% gluteraldehyde to your Tetrahymena and ink solution and wait for 1 minute. The glutaraldehyde will fix (kill) the cells, allowing you to observe the Tetrahymena in more detail.
- IN THE HOOD, add 20 μL of the resulting 30 μL mixture to a glass slide and place a cover slip on top. You can now remove the slide from the hood, but be sure to dispose of this slide in the hood when you are done.
- Using a digital camera, take photos of the Tetrahymena at the 0, 5, 10 and 15 minute timepoints. Please refer to the directions in Appendix H for detailed camera instructions.
- Count the number of ink filled vacuoles in 10 individual Tetrahymena at each time point and record this information in your lab notebook.
- Using the micrometer, measure the size of these phagocytic vacuoles inside the Tetrahymena. Measure at least 1 vacuole from at least 5 individual Tetrahymena. Check with your instructor to be sure that you are measuring the correct structure. Did you see them form in the live "Tetrahymena"? Describe the process of phagocytosis in Tetrahymena in your lab notebook.
PART III: Design your own experiment to investigate a variable that affects phagocytosis in Tetrahymena pyriformis
In the next lab, you will have the opportunity to design your own experiment to study a factor that affects phagocytosis in Tetrahymena. The cytoskeleton plays an important role in the architecture of the cell. Microfilaments (composed of actin) are located throughout the cell but they are found predominantly near the plasma membrane where they play a role in cell movement. Microtubules (composed of tubulin) form hollow tubes in the cell and serve as tracks along which materials are transported. They also play a key role in the organization of organelles and they form the mitotic spindle. Many research papers that explore phagocytosis propose that the cytoskeleton, particularly the actin containing elements of the cytoskeleton, are crucial to some or all of the parts of phagocytosis. There seems to be some conflicting evidence addressing the role of the tubulin containing part of the cytoskeleton. You could choose to explore either the role of microfilaments (actin) or microtubules (tubulin) in phagocytosis in Tetrahymena pyriformis. Alternatively, you may choose to investigate whether particle size affects phagocytosis.
Before you leave today, in groups of 4, design your own experiment to answer one of several questions that may have come up from your observations or from your reading about Tetrahymena phagocytosis. Your instructor will explain your options and she/he will direct you to a research paper or other information related to the question that you and your group chooses to explore. The information given should help you figure out appropriate experimental conditions. You should start by answering the Experimental Design Questions provided by your instructor.
After you have answered these questions and talked to your instructor, you can begin to work on your protocol. Create a sharable document with your group including your names, experimental question, and experimental protocol (including showing how you will make all working solutions of reagents) and division of labor within the group. Make sure your protocol is approved by your instructor.
Main Topic Choices:
1.What role does the cytoskeletal element actin (in microfilaments) play in one or more aspects of phagocytosis (either engulfment, phagosome formation, filling, movement or vesicle fusion) in Tetrahymena pyriformis? Study phagocytosis of India ink in the absence and presence of the actin inhibitor cytochalasin B.
2.What role does the cytoskeleton element tubulin (in microtubules) play in some aspect of phagocytosis (either engulfment, phagosome formation, filling, movement or vesicle fusion) in Tetrahymena pyriformis? Study phagocytosis of India ink in the absence and presence of the tubulin inhibitors colchicine or nocodazole.
3.Is phagocytosis in Tetrahymena pyriformis selective or non-selective for particle size (Study phagocytosis of Polybead dyed microspheres that vary in color and/or size OR study phagocytosis of microspheres and India ink)
Stock concentrations of reagents available to you:
In addition to the reagents that you used previously, cytoskeleton inhibitors and Polybead dyed microspheres (different colors and sizes) will be available for this experiment. CAUTION: Wear gloves and exercise caution when handling the inhibitors because they are toxic.
Cytochalasin B 14mM (6.7mg/ml) in DMSO
Colchicine 25mM (10mg/ml) in water
Nocodazole 7mM (2mg/ml) in DMSO
India ink (Speedball, Product #3328, #3338, Statesville, NC), according to the manufacturer, contains (vol./vol.): water (75-85%), carbon black (7-9%), shellac (6-8%), ammonium hydroxide (1-2%), phenol (0.45%) and ethylene glycol (1.6%). The carbon black is dispersed as ~ 20nm carbon particles but it ranges, in the commercial grade used for India ink from 13-75nm or larger when aggregated.
Polybead Dyed Microspheres available:
1.0µm Blue
3.0µm Blue
6.0µm Blue
10.0µm Blue
3.0µm Yellow
6.0µm Yellow
Polybeads are packaged as 2.5% aqueous suspension (water) and will be diluted to a working concentration of 0.08% prior to lab. This concentration approximately equals the working concentration of carbon particles in 1% India ink.
Laboratory Cleanup
- Place used microcentrifuge tubes and tips that contain gluteraldehyde WITH GLYCINE in autoclave bags taped above your bench.
- Any microcentrifuge tubes containing glutaraldehyde WITHOUT GLYCINE should be disposed of in the hood.
- Put all used microscope slides and cover slips in the glass disposal box.
- Proper storage of your microscope: Clean the objective lenses of your microscope, beginning with the lowest power (4x) and proceeding to the highest. Use lens tissue (NOT Kimwipes®). Make sure that there there is NO oil on any lens. Rotate the 4x objective lens into the locked viewing position. The binocular head must be rotated into the storage position, to protect the ocular lenses from damage. Loosen the setscrew on the right, rotate the head 180°, then retighten the screw. Turn off the microscope light. Return the microscope to the cabinet.
Assignments
The following assignment consists of several parts. This assignment must be done individually.
- Generate two figures from your ink experiment. The first figure should be a composite photomicrograph of Tetrahymena at different time points during your experiment and the second figure should contain a graph of the analyzed data. Both figures should include figure legends. In the Resources section of the lab wiki, find the Guide to Scientific Writing. Refer to the sections on figures and legends for help. Digital image processing can be done with a variety software programs including Powerpoint and iPhoto. If you have a Mac, you might prefer to use your already installed iPhoto application to edit your images.
- What experimental question were you able to address with the data presented in your figures? Please limit this answer to one concise sentence.
- What can you conclude from the data presented in your figures? Please limit this answer to one short paragraph.
- Next week, you will be investigating a variable that affects phagocytosis in Tetrahymena. List your variable and then discuss why you think your particular variable may play a role in Tetrahymena phagocytosis. Limit your response to one paragraph. You must refer to at least one primary article in your response. This article can be the same one suggested by your instructor during lab today. Be sure to properly cite any sources you use to answer this question.