CH391L/S12/Selectablegeneticmarkers: Difference between revisions
No edit summary |
No edit summary |
||
| Line 23: | Line 23: | ||
Oftentimes, researchers find themselves working not with E. coli or other bacteria, but with plant organisms that are unaffected by antibiotics. In these instances, antibiotic resistance is replaced with herbicidal resistance. While the overall process remains essentially the same, herbicide resistance falls under a different category of selectable genetic markers. | Oftentimes, researchers find themselves working not with E. coli or other bacteria, but with plant organisms that are unaffected by antibiotics. In these instances, antibiotic resistance is replaced with herbicidal resistance. While the overall process remains essentially the same, herbicide resistance falls under a different category of selectable genetic markers. | ||
One of the most common forms of herbicide resistance found in the world is glyphosate resistance. The Monsanto corporation introduced glyphosate resistance into soybeans in 1996, and provides an example of the commercial application of selectable genetic markers. Since then, Monsanto has incorporated glyphosate resistance into other plants such as canola, corn, and alfalfa. Approximately 50% of all agricultural land in the United States is now occupied by these variants, attesting to the power of selectable genetic markers<cite>Owen2010</cite>. | One of the most common forms of herbicide resistance found in the world is glyphosate resistance. Glyphosate, a common herbicide found especially in Roundup, is a competitor of the enzyme 5-enolpyruvoyl-shikimate-3-phosphate synthetase (EPSPS). The herbicide acts as a transition state analog, binding readily to EPSPS and thus inhibiting the Shikimate pathway. Monsanto introduced glyphosate resistance by first isolating a variant of EPSPS from Agrobacterium (a gram-negative bacteria) strain CP4 in the 1980s, with the unique feature of not being inhibited by glyphosate<cite>Funke2006</cite>. The Monsanto corporation introduced glyphosate resistance into soybeans in 1996, and provides an excellent example of the commercial application of selectable genetic markers. Since then, Monsanto has incorporated glyphosate resistance into other plants such as canola, corn, and alfalfa. Approximately 50% of all agricultural land in the United States is now occupied by these variants, attesting to the power of selectable genetic markers<cite>Owen2010</cite>. | ||
Selectable genetic markers for plants are not always in the form of herbicide resistance. For instance, researchers at China's Agricultural University were able to express the rstB gene in tobacco, which confers upon the plant greater tolerance to salt concentration. <i>Zhang et al.</i> was able to | Selectable genetic markers for plants are not always in the form of herbicide resistance. For instance, researchers at China's Agricultural University were able to express the rstB gene in tobacco, which confers upon the plant greater tolerance to salt concentration. <i>Zhang et al.</i> was able to achieve approximately 80% selection efficiency using salt concentrations at 170mM, proving remarkable success in using a selectable genetic marker other than herbicide resistance<cite>Zhang2008</cite>. | ||
==TO ADD: Auxotrophic== | |||
===Other=== | ===Other=== | ||
| Line 52: | Line 54: | ||
Selectable genetic markers can also be used to investigate protein activity. Research performed at the University of Washington used GFP coupled with inteins to splice GFP into other proteins with greater than 96% efficiency. The GFP proved harmless to the intein's activity, and also allowed the researchers to analyze the effectiveness of the inteins themselves<cite>Ramsden2011</cite>. | Selectable genetic markers can also be used to investigate protein activity. Research performed at the University of Washington used GFP coupled with inteins to splice GFP into other proteins with greater than 96% efficiency. The GFP proved harmless to the intein's activity, and also allowed the researchers to analyze the effectiveness of the inteins themselves<cite>Ramsden2011</cite>. | ||
TO ADD: Neat story about GFP's discoverer | |||
| Line 102: | Line 106: | ||
#Ramsden2011 pmid=21708017 | #Ramsden2011 pmid=21708017 | ||
//Use of inteins with GFP to introduce selectable markers into proteins. | //Use of inteins with GFP to introduce selectable markers into proteins. | ||
#Funke2006 pmid=16916934 | |||
//Molecular basis of glyphosate resistance. | |||
</biblio> | </biblio> | ||
Revision as of 19:31, 22 February 2012
Selectable Markers Overview
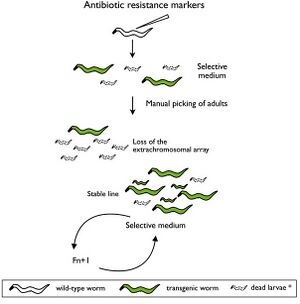
Selectable genetic markers are exogenous genes that are introduced into a cell, conferring a previously absent selective advantage. These markers are primarily used to "mark" the successful ligation of DNA into a plasmid and subsequent transformation into a cell. Oftentimes, selectable markers are accompanied by other exogenous genes that is the primary gene of interest; the marker simply serves to distinguish between successful transformations, and unaltered wild-type cells.
It is not atypical to witness transformation efficiencies as low as .05%, making it difficult to pick correct cellular colonies without additional techniques. This is where the selectable genetic markers prove their usefulness. For instance, selectable genetic markers can be used to confer ampicillin resistance to E. coli. These newly resistant E. coli can then be grown on culture plates with ampicillin, allowing only E.coli with successfully transformed DNA to proliferate.
In addition to selectable genetic markers are screenable genetic markers. Screenable genetic markers function in a similar manner in that they are exogenous genes that are transformed into a cell; however, they do not confer any new sort of resistance to the cell. Instead, they cause the cell to respond differently to environmental conditions in such a way as to distinguish transformed cells from untransformed cells. This can be useful when determining the transformation efficiency of a cell, or when carefully monitoring the activity of proteins.
Types of Selectable Markers
Antibiotic
In synthetic biology research, the primary forms of selectable markers are antibiotic resistant genes. Because a large portion of research takes place in vivo in E. coli, antibiotic selectable markers can be employed whenever transfecting DNA in order to distinguish wild-type cells from successfully transfected ones. If ligating more than one gene of interest into a plasmid for transfection into E. coli, it is often beneficial to employ multiple antibiotic markers to ensure that both genes are present in resultant colonies.
Common types of antibiotics used include ampicillin, tetracycline, chloramphenicol, and the many -mycins, including kanamycin. A large range of antibiotic resistances are used as genetic markers. Because of this, each antibiotic resistance is often referred to by a three letter acronym, such as Amp, Tet, Chl, Cam and Kan. Plates containing these antibiotics can be made en mass, and used to grow appropriate cultures of transformed E. coli.
Antibiotic markers are the most popular form of selectable genetic markers. As such, the field is quite large and constantly expanding in order to meet research needs. For instance, Poggi et al. recognized the mutation of antibiotic resistance towards gentamicin, kanamycin, streptomycin, and spectinomycin in leptospiral pathogens. The group was able to develop a cassette that included two antibiotic markers, along with a new gentamicin marker. Using multiple antibiotic markers greatly reduces the chance of background colonies that have spontaneously developed antibiotic resistance[2].
Herbicidal
Oftentimes, researchers find themselves working not with E. coli or other bacteria, but with plant organisms that are unaffected by antibiotics. In these instances, antibiotic resistance is replaced with herbicidal resistance. While the overall process remains essentially the same, herbicide resistance falls under a different category of selectable genetic markers.
One of the most common forms of herbicide resistance found in the world is glyphosate resistance. Glyphosate, a common herbicide found especially in Roundup, is a competitor of the enzyme 5-enolpyruvoyl-shikimate-3-phosphate synthetase (EPSPS). The herbicide acts as a transition state analog, binding readily to EPSPS and thus inhibiting the Shikimate pathway. Monsanto introduced glyphosate resistance by first isolating a variant of EPSPS from Agrobacterium (a gram-negative bacteria) strain CP4 in the 1980s, with the unique feature of not being inhibited by glyphosate[3]. The Monsanto corporation introduced glyphosate resistance into soybeans in 1996, and provides an excellent example of the commercial application of selectable genetic markers. Since then, Monsanto has incorporated glyphosate resistance into other plants such as canola, corn, and alfalfa. Approximately 50% of all agricultural land in the United States is now occupied by these variants, attesting to the power of selectable genetic markers[4].
Selectable genetic markers for plants are not always in the form of herbicide resistance. For instance, researchers at China's Agricultural University were able to express the rstB gene in tobacco, which confers upon the plant greater tolerance to salt concentration. Zhang et al. was able to achieve approximately 80% selection efficiency using salt concentrations at 170mM, proving remarkable success in using a selectable genetic marker other than herbicide resistance[5].
TO ADD: Auxotrophic
Other
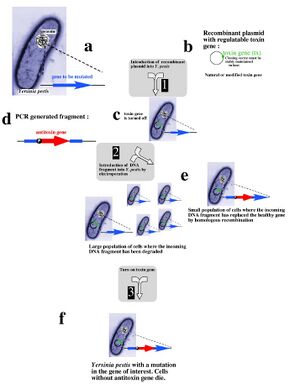
Recent research into selectable genetic markers has looked into pathways that avoid employing antibiotic and herbicidal resistance. This is due to rising concern over "wild" strains of bacteria or plants developing antibiotic or herbicidal resistance and proliferating rapidly in nature. Even in a laboratory environment, avoiding the resistance approach towards selectable markers can prove beneficial.
A novel approach towards selectable markers was developed in Lawrence Livermore National Laboratory, which employes a toxin/antitoxin combination of genes as a marker. The process, summarized in the figure to the left, effectively avoids the need to grow antibiotic resistant bacterial cultures on an antibiotic plate. An inducible zeta-toxin group of proteins is first introduced into an E. coli strain. A DNA strand of interest containing an zeta-antitoxin group is then transformed into the E. coli, and the entire culture is grown. The zeta-toxin group is then induced, killing off all E. coli that does not contain the antitoxin group. Besides for triggering the zeta-toxin group, no outside influence is required to select for the desired cells[6].
Types of Screening
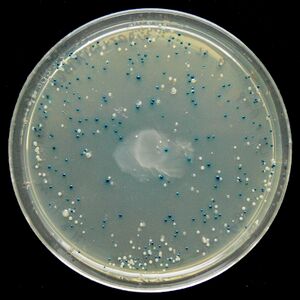
Blue/White Screening
Blue/White Screening is commonly used in E. coli transformations. In this screening, cells are grown on agar plates in the presence of X-gal and IPTG to test for the presence of β-galactosidase enzyme. In the M15 strain of E. coli, part of the lacZ gene is deleted, removing the cell's ability to produce β-galactosidase. However, when transfected with a plasmid containing a lacZα domain, such as pUC19, the gene becomes operable and the cell produces β-galactosidase. It is possible to create a successful transformation in which β-galactosidase is not produced by inserting DNA into the lacZα domain. This is particularly useful to check for successful ligations. Successful ligations will not produce β-galactosidase, while unsuccessful ligations will.
X-gal, while normally colorless (i.e. white), will readily hydrolyze in the presence of β-galactosidase into a compound with a sharp blue color. Therefore, colonies with successfully transformed cells with the desired DNA will grow white, while background colonies will grow blue.
Green Fluorescent Protein Screening
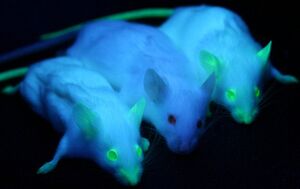
Green Fluorescent Protein, or GFP, was first isolated from the crystal jellyfish Aequorea victoria in the 1960s. In 1994, GFP was successfully cloned[8], allowing researchers to use the protein as a screenable marker for the first time. Virtually harmless in live cells, GFP has the unique pheotype of glowing bright green under ultraviolet light. GFP functions entirely of its own accord, and requires no exogenous material besides ionizing radiation in order to fluoresce. This allows GFP to be used as a marker accompanying transfected DNA, and has been used extensively in academia.
In 2011, GFP was used to create an in vivo mammary model to investigate tumorigenesis in mice. Tumor cells were introduced into the mice, accompanied with GFP as a screenable marker. As the mice tumors proliferated, so did GFP. This allowed for easy differentiate between tumors and stroma cells, greatly aiding cancer researchers[7].
Selectable genetic markers can also be used to investigate protein activity. Research performed at the University of Washington used GFP coupled with inteins to splice GFP into other proteins with greater than 96% efficiency. The GFP proved harmless to the intein's activity, and also allowed the researchers to analyze the effectiveness of the inteins themselves[9].
TO ADD: Neat story about GFP's discoverer
Artificial Selection
Selectable markers have a remarkably relevant role in industrial applications. Because of their ability to distinguish cells from one another, selectable markers are an essential tool for artificial selection. While artificial selection of organisms is possible without the use of selectable markers, the process is significantly shorter with their use.
Artificial selection is a special instance in which selectable markers are often the desired gene to be introduced into a cell. For instance, rice has been transfected with a plethora of resistances using selectable markers. Glycopeptide binding protein, dihydrofolate reductase, and hygromycin phosphotransferase have all been introduced into rice, conferring resistance to bleomycin and pheomycin, methotrexate, and hygromycin B respectively. This allows farmers to use herbicides select for only rice with these markers, while eliminating the majority of invasive species[10].
Issues
As mentioned earlier, current research into genetic markers involves looking at alternatives to antibiotic and herbicidal resistance. Both the commercial and research sectors have reason to adopt alternative techniques. In the commercial sector, there is the omnipresent public fear of growing and consuming "genetically modified crops". In the research sector, new techniques can decrease costs, time, labor, and error rates (it is always possible that wild-type cultures can grow under antibiotic/herbicidal conditions). Either sector would also be held responsible by governments should their modified organisms escape into the environment and proliferate.
Example: Proliferation of glyphosate resistance
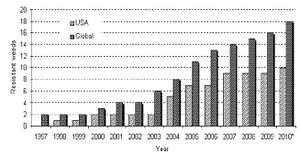
Since Monsanto's introduction of glyphosate resistance in 1996, there has been a growing concern over the affects of its widespread use. Glyphosate resistance is used primarily in Monsanto's Roundup Ready™ plants to work in tandem with Monsanto's Roundup™ herbicide, whose key active ingredient is glyphosate. Roundup has been used commercially since 1976. However, Roundup's true success did not truly begin until the development of Roundup Ready selective genetic markers.
It was at this time that an explosion in glyphosate resistant weeds began in the environment. Originally, there were no none plants natively resistant to glyphosate. Even in 1996, 30 years after the introduction of Roundup, only 2 weeds worldwide had developed glyphosate resistance. However, as shown in the figure to the right, this number increased to 19 weeds by 2010[4]. The correlation between the introduction of commercial glyphosate resistance and the mutation of invasive weeds is substantial, and raised public concerns.
In 2006, after hearing a case brought against Monsanto by a coalition of farmers and environmental groups, the US District Court of Northern California ruled against Monsanto and issued an injunction on all Roundup Ready alfalfa sales. An appeal to the Ninth Circuit Court of Appeals upheld the District Court's ruling. It was not until a Supreme Court hearing in January of 2010 that the decision was reversed and the injunction lifted.
References
- Giordano-Santini R and Dupuy D. Selectable genetic markers for nematode transgenesis. Cell Mol Life Sci. 2011 Jun;68(11):1917-27. DOI:10.1007/s00018-011-0670-1 |
Review article about selectable genetic markers as used in nematodes. Relatively new field for nematodes, possible due to the completion of the Caenorhabditis elegans genome.
- Poggi D, Oliveira de Giuseppe P, and Picardeau M. Antibiotic resistance markers for genetic manipulations of Leptospira spp. Appl Environ Microbiol. 2010 Jul;76(14):4882-5. DOI:10.1128/AEM.00775-10 |
Multiple antibiotic resistances as selectable genetic markers in Leptospira.
- Funke T, Han H, Healy-Fried ML, Fischer M, and Schönbrunn E. Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13010-5. DOI:10.1073/pnas.0603638103 |
Molecular basis of glyphosate resistance.
-
Report by Professor Micheal Owen from Iowa State University on glyphosate resistance for the U.S. House of representatives
-
Zhang WJ, Yang SS, Shen XY, Jin YS, Zhao HJ and Wang T
Salt tolerance as a selectable genetic marker.
-
D. Parsons, M. Tolmasky, P. Chain and B. W. Segelke
Report by the Lawrence Livermore National Laboratory on a new system for selectable markers
- Moen I, Jevne C, Wang J, Kalland KH, Chekenya M, Akslen LA, Sleire L, Enger PØ, Reed RK, Øyan AM, and Stuhr LE. Gene expression in tumor cells and stroma in dsRed 4T1 tumors in eGFP-expressing mice with and without enhanced oxygenation. BMC Cancer. 2012 Jan 17;12:21. DOI:10.1186/1471-2407-12-21 |
Observes tumor growth in mice by introducing GFP into the mice.
- Chalfie M, Tu Y, Euskirchen G, Ward WW, and Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994 Feb 11;263(5148):802-5. DOI:10.1126/science.8303295 |
The first instance of using GFP as a marker.
- Ramsden R, Arms L, Davis TN, and Muller EG. An intein with genetically selectable markers provides a new approach to internally label proteins with GFP. BMC Biotechnol. 2011 Jun 27;11:71. DOI:10.1186/1472-6750-11-71 |
Use of inteins with GFP to introduce selectable markers into proteins.
-
Twyman RM, Stroger E, Kohli A, Capell T and Christou P
Various genetic markers used for artificial selection of rice crops.
- Goodwin JL, Pastori GM, Davey MR, and Jones HD. Selectable markers: antibiotic and herbicide resistance. Methods Mol Biol. 2005;286:191-202. DOI:10.1385/1-59259-827-7:191 |
Herbicide selectable genetic markers in wheat.