BioMicroCenter:Microarrays: Difference between revisions
| Line 72: | Line 72: | ||
[http://www.affymetrix.com AFFYMETRIX] arrays are very high density arrays of 25mer oligonucleotides built by lithography. An Affymetrix probeset is composed of numerous neighboring perfect match and mismatch probes that are combined and used in the calculation of intensity and relative expression. Affymetrix arrays are the most common arrays used in genomics right now and have a rich bioinformatic architecture that allows for a large number of analyses. While expression arrays are the most common, Affymetrix also creates arrays for SNP calling and ChIP-chip. Affymetrix expression arrays come in two flavors: 3' and exon. 3' arrays use oligo dT priming while exon arrays amplify from the whole RNA using random primers. Affymetrix arrays are one-color arrays and use biotinylated nucleotides in the cRNA/cDNA to recruit HRP-peroxidase to the probes. Affymetrix arrays come as single arrays from a broad list of species. The BioMicro Center has a hybridization oven, two 450 fluidics devices and a 7G scanner for handling Affymetrix arrays. | [http://www.affymetrix.com AFFYMETRIX] arrays are very high density arrays of 25mer oligonucleotides built by lithography. An Affymetrix probeset is composed of numerous neighboring perfect match and mismatch probes that are combined and used in the calculation of intensity and relative expression. Affymetrix arrays are the most common arrays used in genomics right now and have a rich bioinformatic architecture that allows for a large number of analyses. While expression arrays are the most common, Affymetrix also creates arrays for SNP calling and ChIP-chip. Affymetrix expression arrays come in two flavors: 3' and exon. 3' arrays use oligo dT priming while exon arrays amplify from the whole RNA using random primers. Affymetrix arrays are one-color arrays and use biotinylated nucleotides in the cRNA/cDNA to recruit HRP-peroxidase to the probes. Affymetrix arrays come as single arrays from a broad list of species. The BioMicro Center has a hybridization oven, two 450 fluidics devices and a 7G scanner for handling Affymetrix arrays. | ||
[http://www.agilent.com AGILENT] arrays are lower density arrays then Affymetrix, but have much longer oligos (60mers) and are printed on glass slides using dot-matrix printing technologies. Because this method does not require the creation of metal plates, novel microarray designs cost no more then designs created by Agilent. Agilent arrays can be used for a variety of assays including gene expression, ChIP-chip, miRNA hybridization, and CGH. Our Agilent scanner was recently upgraded and can handle Agilent arrays of up to 1 million probes per slide (Donated by the Bell lab). Because of the lower probe density and the longer oligos used, each probe is evaluated separately. [[Image: | [http://www.agilent.com AGILENT] arrays are lower density arrays then Affymetrix, but have much longer oligos (60mers) and are printed on glass slides using dot-matrix printing technologies. Because this method does not require the creation of metal plates, novel microarray designs cost no more then designs created by Agilent. Agilent arrays can be used for a variety of assays including gene expression, ChIP-chip, miRNA hybridization, and CGH. Our Agilent scanner was recently upgraded and can handle Agilent arrays of up to 1 million probes per slide (Donated by the Bell lab). Because of the lower probe density and the longer oligos used, each probe is evaluated separately. [[Image:Multiple_arrays.jpg|right]] Each glass slide can contain multiple identical arrays (2, 4, or 8), reducing the cost per hybridization, often below the level of Affymetrix arrays. Typical mammalian expression arrays are in a 4x44k or 8x60k format where each slide contains four or eight identical arrays that are separable by a gasket slide. Agilent arrays can be hybridized either as one-color or two-color using Cy5 and Cy3 labeled cRNA/cDNA. We do not currently maintain a stock of Agilent arrays but do have a significant discount on the purchase of arrays. | ||
Please note that we have upgraded our Agilent DNA microarray scanner to the new scanning system enabled by SureScan High-Resolution Technology on April 2011. The new scanner is enable scanning of with 2, 3, 5 or 10 micron resolution glass slide microarrays; identifying weaker signals and preventing feature saturation; speeding up scan times; providing increased sensitivity and precision by dynamic autofocus, regardless of glass aberrations. That allows scanning of Agilent’s newest 1 million feature arrays. | Please note that we have upgraded our Agilent DNA microarray scanner to the new scanning system enabled by SureScan High-Resolution Technology on April 2011. The new scanner is enable scanning of with 2, 3, 5 or 10 micron resolution glass slide microarrays; identifying weaker signals and preventing feature saturation; speeding up scan times; providing increased sensitivity and precision by dynamic autofocus, regardless of glass aberrations. That allows scanning of Agilent’s newest 1 million feature arrays. | ||
Revision as of 17:46, 28 June 2011
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |

The MIT BioMicro Center has extensive experience with several different types of microarray samples and platforms. Microarray labeling and hybridization are typically performed by BioMicro Center technicians, however our scanners, hybridization ovens and wash stations are available by request.
BioMicro Center Services
We are currently set up to primarily handle expression analysis. Our service includes:
- Evaluation of sample quality using the Agilent BioAnalyzer 2100
- Sample labeling and clean up
- Array Hybridization
- Data extraction and normalization
We also have discounts with different vendors and can help you purchase your arrays.
SAMPLE TYPES
The BioMicro Center has extensive experience handling samples for expression analysis.
| Eukaryote | Min.RNA | Min.Vol. | Platforms | Notes |
|---|---|---|---|---|
| Prokaryote | 1 μg | 6 μL | Affy or Agilent | labeling using Ambion for Affy arrays |
| 3' Array (std) | 50ng / 100pg | 6 μL | Affy or Agilent | labeling using NuGEN for Affy arrays. |
| Exon Array | 50ng / 100pg | 6 μL | Affymetrix | labeling using NuGEN for Affy arrays |
| CGH | 3ug (DNA) | 6 μL | Agilent | labeling using Agilent's CGH kit |
| small RNA | 1ug | 6 μL | Agilent | labeling using Agilent's miRNA arrays |
SAMPLE SUBMISSION
We encourage researchers to contact us several days in advance of sample submission to allow us lead time to purchase any specialized Affymetrix or Agilent arrays. While we keep a few arrays in stock, most arrays are ordered on an as needed basis. A list of some of the available arrays and their prices can be found in this file. Information about Affymetrix arrays is available at http://www.affymetrix.com/products_services/index.affx
All submissions must be accompanied by a Affymetrix or Agilent Submission Form.
TURN AROUND
The BioMicro Center will turn around your samples as quickly as possible. We are currently exploring methods of increasing our sample throughput to reduce turn around time. Currently the average turn around time is 1-2 weeks.
ARRAY SELECTION
There are several considerations to take into account in selecting your microarray. We provide support for multiple array platforms from two different manufacturers (Agilent and Affymetrix). Additionally, multiple labeling methods are available for each platform. Each labeling method has different sample requirements and multiple labeling methods should NOT be combined in doing post-hybridization analysis.
Agilent vs. Affymetrix
AFFYMETRIX arrays are very high density arrays of 25mer oligonucleotides built by lithography. An Affymetrix probeset is composed of numerous neighboring perfect match and mismatch probes that are combined and used in the calculation of intensity and relative expression. Affymetrix arrays are the most common arrays used in genomics right now and have a rich bioinformatic architecture that allows for a large number of analyses. While expression arrays are the most common, Affymetrix also creates arrays for SNP calling and ChIP-chip. Affymetrix expression arrays come in two flavors: 3' and exon. 3' arrays use oligo dT priming while exon arrays amplify from the whole RNA using random primers. Affymetrix arrays are one-color arrays and use biotinylated nucleotides in the cRNA/cDNA to recruit HRP-peroxidase to the probes. Affymetrix arrays come as single arrays from a broad list of species. The BioMicro Center has a hybridization oven, two 450 fluidics devices and a 7G scanner for handling Affymetrix arrays.
AGILENT arrays are lower density arrays then Affymetrix, but have much longer oligos (60mers) and are printed on glass slides using dot-matrix printing technologies. Because this method does not require the creation of metal plates, novel microarray designs cost no more then designs created by Agilent. Agilent arrays can be used for a variety of assays including gene expression, ChIP-chip, miRNA hybridization, and CGH. Our Agilent scanner was recently upgraded and can handle Agilent arrays of up to 1 million probes per slide (Donated by the Bell lab). Because of the lower probe density and the longer oligos used, each probe is evaluated separately.
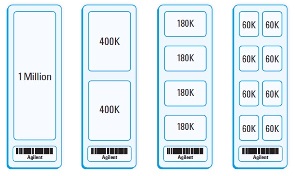
Each glass slide can contain multiple identical arrays (2, 4, or 8), reducing the cost per hybridization, often below the level of Affymetrix arrays. Typical mammalian expression arrays are in a 4x44k or 8x60k format where each slide contains four or eight identical arrays that are separable by a gasket slide. Agilent arrays can be hybridized either as one-color or two-color using Cy5 and Cy3 labeled cRNA/cDNA. We do not currently maintain a stock of Agilent arrays but do have a significant discount on the purchase of arrays.
Please note that we have upgraded our Agilent DNA microarray scanner to the new scanning system enabled by SureScan High-Resolution Technology on April 2011. The new scanner is enable scanning of with 2, 3, 5 or 10 micron resolution glass slide microarrays; identifying weaker signals and preventing feature saturation; speeding up scan times; providing increased sensitivity and precision by dynamic autofocus, regardless of glass aberrations. That allows scanning of Agilent’s newest 1 million feature arrays.
As of July 2011, on a per sample basis, Agilent arrays cost considerably less then Affymetrix Genechips (under $400 per array), but must be processed as whole slides.
Please note that the BioMicro Center no longer supports self printed arrays. Custom oligonuclotide arrays are available from Agilent at the same cost as their commercial arrays using their eArray system (we can help you through the process of designing arrays). For anyone requiring self printed arrays, we encourage you to contact Tom Volkert at the Whitehead Institute CGT.
Labeling Methods
Labeling Kits

Affymetrix retired their old 3' and Exon Array kits recently. BMC evaluated several new technologies to replace the kits. The newer kits are faster and require less starting material. Based on the type of protocol, we have chosen different kits for amplifying and labeling the RNA.Information on the evaluation can be found HERE.. More information about some of our labeling protocols is below.
- Prokaryote arrays: For both Agilent and Affymetrix, we have had success using the [Ambion MessageAmp II-Bacteria kits. These kits require 100ng of total RNA to start with. The RNA is then polyadenylated. The polyA RNA is then reverse transcribed with a T7 containing primer and 2nd strand of DNA is created. The T7 primer is then used to make antisense cRNA incorporating biotinylated or Cy3/Cy5 nucleotides which are hybridized to the arrays. We can use the old Affymetrix labeling system by request.
- Affymetrix 3' arrays | Agilent Arrays (option1): For standard Affymtrix arrays and for some Agilent arrays we have been using the NuGEN Applause system. This system uses 50-100ng of total RNA as input. A unique feature of the NuGEN kits is that they use cDNA for hybridization instead of cRNA. Several studies have found increased specificity with the NuGEN system(eg. Eklund et al., Nat.Biotech. 2006). Preliminary tests were performed using samples from the White lab and analyzed in collaboration with Charlie Whittaker at the Koch Institute Bioinformatics and Computing Core Facility. The NuGEN kits showed significantly higher sensitivity then the Affymetrix kits coupled with decreased levels of background hybridization. All of the kits showed highly similar changes in expression between two samples, however the absolute level of detected transcripts were quite different. NuGEN kits can be adapted for Agilent arrays much the same way the Ambion prokaryote kits can.
- Affymetrix Exon Arrays : For whole transcriptome analysis, we have two kits we are currently evaluating. We have the most experience with the NuGEN wt-ovation Exon kit, which works much the same way as the 3' kit does. We are also evaluating the Ambion exon kit which is recommended by Affymetrix. Standard kits for both can handle samples over 50ng. The NuGEN kit is unique in that it can handle samples down to 500pg using their Pico kit. NuGEN also sells a [single cell preparation kit. Please contact Stuart Levine if you are interested in trying out these technologies.
We DO NOT RECOMMEND mixing labeling types within an experiment as we have observed large differences between the absolute intensities of signals between labeling methods.
QUALITY CONTROL
The BioMicro Center employs several quality controls as part of microarray analysis
RNA
All RNA samples are run on the Agilent BioAnalyzer prior to labeling. The BioAnalyzer results are used to determine the concentration of the RNA and the quality of the RNA based on their RNA Integrity Number (RIN) score.
Affymetrix Arrays
After scanning, 3' Affymetrix arrays are processed through a number of quality control steps to confirm the quality of the array. These steps create quality control plots that we distribute to the user.
QC PLOTS
In all cases, the plots are guidelines but not absolutes. Differences in the quality of starting RNA or source of the RNA can have a major impact on the plots. Biological replicates should behave similarly and we focus on changes where we find that is not true.
The RNA Degradation plot measure the relative intensity of probes at the 5' and 3' end of the average gene. Differences in the reverse transcription step can hive significant effects on the outcome. The precise slope of the line is not as important as having the lines have similar slopes.
Normalized Unscaled Standard Error (NUSE), provides a measure of relative chip quality derived from the residuals from the RMA model. NUSE scores above 1.05 or disconnected from the others are generally (though not always) considered poor.
The Affy QC pdf contains a number of metrics including box plots and histograms of intensity, QC scores for spike in controls and interarray correlation.
Fianlly, we use gcRMA for normalization. gcRMA normalized data is included in the data we distribute.
References
These plots are generated using a number of packages from bioconductor including:
- affy:Gautier, L., Cope, L., Bolstad, B. M., and Irizarry, R. A. 2004. affy---analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 3 (Feb. 2004), 307-315.
- affyQCReport
- affyPLM: Brettschneider J, Collin F, Bolstad BM, and Speed TP. (2007) Quality assessment for short oligonucleotide arrays. Technometrics.
- LPE: Nitin Jain, Michael O'Connell and Jae K. Lee. Includes R source code contributed by HyungJun Cho <hcho@virginia.edu> (). LPE: Methods for analyzing microarray data using Local Pooled Error (LPE) method. R package version 1.20.0. http://www.r-project.org,
Bioconductor reference: Bioconductor: Open software development for computational biology and bioinformatics R. Gentleman, V. J. Carey, D. M. Bates, B.Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, and others 2004, Genome Biology, Vol. 5, R80
gcRMA: Jean(ZHIJIN) Wu and Rafael Irizarry with contributions from James MacDonald Jeff Gentry (). gcrma: Background Adjustment Using Sequence Information. R package version 2.18.1.
Agilent Arrays
- - To Be Added - -
DATA ANALYSIS
Affymetrix Data Analysis
BioMicro Center uses Affymetrix Expression Console and GCRMA for analysis of Affymetrix data. Several files for each sample will be provided to the user:
- .exp: Experiment information file
- .dat: The image of the scanned probe array.
- .cel: Cell Intensity File derived from .dat file.
- .chp: Output file generated from the analysis of a probe array.
- .rpt: Report File generated.
- .txt: Text File is the same information as the .chp file in text format.
Additional data analysis help is available on a case by case basis. Please contact Stuart Levine if you need additional help with your analysis.
Agilent Data Analysis
Agilent Scanner is available to members of the MIT Community for $50 per scan. We are currently running Feature extractor version 9.2. For questions about equipment, contact Manlin Luo After you have completed your scan, please make sure to sign the scanner record form, which is near the equipment and to remove your files from the computer. CD-Rs are available if needed.
LINKS
You will be notified by email that your data is ready. At that time please pick up any leftover samples and arrays belonging to you. Samples will be held at the facility at -80C for 60 days after your data is generated. Information on downloading your data is in the link above.
