Wilke: Difference between revisions
No edit summary |
|||
| Line 11: | Line 11: | ||
== 04/22/2011—Contact Networks Shape Parasite Evolutionary Trees == | == 04/22/2011—Contact Networks Shape Parasite Evolutionary Trees == | ||
[[Image:Wilke_newsicon_04222011.png|left|150px]] The inference of population dynamics (such as the number of infected individuals as a function of time) from molecular sequence data is becoming an important new method for the surveillance of infectious diseases. We have examined how heterogeneity in host contacts shapes the genealogies of parasitic agents. We find that contact heterogeneity can have a strong effect on how the structure of genealogies reflects epidemiologically relevant quantities such as the proportion of a population that is infected. Contact heterogeneity also can increase the number of sequence isolates required to estimate these quantities over the course of an epidemic. Our results suggest that data about contact-network structure will be required in addition to sequence data for accurate estimation of a parasitic agent's genealogy. This work is published in a special issue of the journal Perspectives on Infectious Diseases focused on [http://www.hindawi.com/journals/ipid/2011/ | [[Image:Wilke_newsicon_04222011.png|left|150px]] The inference of population dynamics (such as the number of infected individuals as a function of time) from molecular sequence data is becoming an important new method for the surveillance of infectious diseases. We have examined how heterogeneity in host contacts shapes the genealogies of parasitic agents. We find that contact heterogeneity can have a strong effect on how the structure of genealogies reflects epidemiologically relevant quantities such as the proportion of a population that is infected. Contact heterogeneity also can increase the number of sequence isolates required to estimate these quantities over the course of an epidemic. Our results suggest that data about contact-network structure will be required in addition to sequence data for accurate estimation of a parasitic agent's genealogy. This work is published in a special issue of the journal Perspectives on Infectious Diseases focused on [http://www.hindawi.com/journals/ipid/2011/238743/ network perspectives on infectious disease dynamics]. | ||
== 03/04/2010—Universal trend of reduced RNA stability near translation-initiation site == | == 03/04/2010—Universal trend of reduced RNA stability near translation-initiation site == | ||
Revision as of 21:05, 22 April 2011
The page you are looking at is kept for archival purposes and will not be further updated.
The Wilke lab carries out research in computational evolutionary biology, bioinformatics, and biostatistics. All our research is theoretical or computational, but we frequently collaborate with experimental groups. Much of our research focuses on molecular evolution, in particular on (i) the evolution of viruses and (ii) biophysical mechanisms of protein evolution. Other areas of interest are theoretical population genetics, epidemiology, and immunology.
To see what we are currently working on, check out our recent publications. Interested in joining? Click here.
Recent News
04/22/2011—Contact Networks Shape Parasite Evolutionary Trees
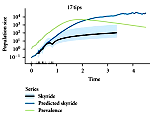
The inference of population dynamics (such as the number of infected individuals as a function of time) from molecular sequence data is becoming an important new method for the surveillance of infectious diseases. We have examined how heterogeneity in host contacts shapes the genealogies of parasitic agents. We find that contact heterogeneity can have a strong effect on how the structure of genealogies reflects epidemiologically relevant quantities such as the proportion of a population that is infected. Contact heterogeneity also can increase the number of sequence isolates required to estimate these quantities over the course of an epidemic. Our results suggest that data about contact-network structure will be required in addition to sequence data for accurate estimation of a parasitic agent's genealogy. This work is published in a special issue of the journal Perspectives on Infectious Diseases focused on network perspectives on infectious disease dynamics.
03/04/2010—Universal trend of reduced RNA stability near translation-initiation site
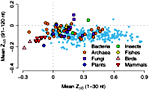
In an analysis of 340 complete genomes from bacteria, archaea, and eukaryotes including fungi, plants, insects, fish, birds, and mammals, we have found a universal trend for reduced RNA stability near the translation-initiation site. With few exceptions, the secondary structure of mRNAs is less stable than expected in the first 30-50 nucleotides downstream from the start codon, but is more stable than expected further downstream. The effect is strongly correlated with genomic GC content — the higher GC the stronger the destabilization effect — and, in prokaryotes, with growth temperature — the lower the optimal growth temperature, the stronger the destabilization effect. These observations are consistent with a thermodynamic hypothesis that stable RNA secondary structure near the start codon can interfere with efficient translation initiation. This work is published in the February issue of PLoS Computational Biology.
02/24/2010—NSF funds BEACON, a Science and Technology Center in Evolutionary Biology

The Wilke lab is part of a $25 million, multi-university center that will study evolution in action in natural and artificial settings. The center is called BEACON, "Bio/computational Evolution in Action CONsortium." It will be headquartered at Michigan State University. Other participating universities are The University of Texas at Austin, the University of Washington, the University of Idaho and North Carolina A&T State University.
Read the UT press release here, or go to the BEACON web-site here.
01/05/2010—New York Times article on lethal mutagenesis
In today's issue of the New York Times, Carl Zimmer discusses the prospects and challenges of combating viruses with lethal mutagenesis. The article features some of the work done in the Wilke lab as well as work done by our colleagues and collaborators in the Bull lab.
09/16/2009—Novel source of HIV-1 viremia in patients on HAART
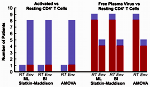
Even though highly active antiretroviral therapy (HAART) can reduce HIV-1 virus load to clinically undetectable levels, the virus never completely disappears and ultrasensitive assays can detect small quantities of virus in all patients on HAART. The exact origin of this virus is unknown. Many researchers assume that it is produced by latently infected CD4+ T cells that reactivate. We analyzed HIV-1 sequences isolated from resting CD4+ T cells, activated CD4+ T cells, and blood plasma using a population-genetics approach. Our analysis showed that sequences from resting and activated CD4+ T cells formed a single population, whereas some of the virus in the blood plasma seemed genetically distinct from the virus in CD4+ T cells. This result shows that circulating CD4+ T cells are not the only source of residual viremia, and it suggests that a novel cellular source may contribute significantly to ongoing virus production under HAART. This research was featured by Science Daily.
06/15/2009—Translational-accuracy selection protects buried and structurally important sites
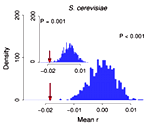
In 1994, Akashi proposed that translationally optimal codons, codons that are translated with relatively low error rate, should preferentially be located at important sites in coding sequences. This signal would be the consequence of translational-accuracy selection, i.e., selection to minimize the amount of non-functional or misfolded protein produced by translation errors. Traditionally, the importance of a site under Akashi's test has been assessed by evolutionary conservation. The July issue of Mol. Biol. Evol. contains a study by Zhou et al. that correlates the location of optimal codons with sites that are important for protein structure. The study finds that there is a tendency of optimal codons to appear at structurally important sites in a wide range of organisms. The study lends further credence to the mistranslation-induced protein-misfolding hypothesis, which argues that much of the selection pressure on coding sequences stems from the toxic effects of mistranslated and misfolded proteins.
05/20/2009—HIV viral-load dynamics under Raltegravir
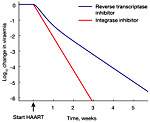
The spectrum of anti-HIV drugs was recently extended by a new class of drugs, the integrase inhibitors. The first drug of this class that received FDA approval is Raltegravir. Clinical data show that when previously untreated patients start treatment on Raltegravir, their viral load declines more rapidly than it does in patients who take instead the reverse-transcriptase inhibitor Efavirenz. This spring, Antiviral Therapy published a modeling study by Sedaghat et al. that discusses the possible mechanisms responsible for this accelerated decline in viral load. The study argues that the accelerated decline is likely not caused by greater antiviral efficiency of Raltegravir compared to Efavirenz. Instead, because Raltegravir acts later in the viral life cycle than Efavirenz, at the beginning of Raltegravir therapy fewer cells have progressed to a state where the drug can not inhibit virus production, and hence the viral load declines faster. The study is a follow-up to a paper published in 2008 in Proc. Natl. Acad. Sci. USA.
References
- Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, and Wilke CO. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J Virol. 2009 Sep;83(17):8470-81. DOI:10.1128/JVI.02568-08 |
- Zhou T, Weems M, and Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol. 2009 Jul;26(7):1571-80. DOI:10.1093/molbev/msp070 |
- Sedaghat AR, Siliciano RF, and Wilke CO. Constraints on the dominant mechanism for HIV viral dynamics in patients on raltegravir. Antivir Ther. 2009;14(2):263-71.
- Sedaghat AR, Dinoso JB, Shen L, Wilke CO, and Siliciano RF. Decay dynamics of HIV-1 depend on the inhibited stages of the viral life cycle. Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4832-7. DOI:10.1073/pnas.0711372105 |