BioMicroCenter:Caliper LabChip GX: Difference between revisions
Manlin Luo (talk | contribs) |
Manlin Luo (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
{{BioMicroCenter}} | {{BioMicroCenter}} | ||
== CALIPER LAB CHIP == | |||
[[Image:Caliper3.jpg|right|200px]] | [[Image:Caliper3.jpg|right|200px]] | ||
| Line 11: | Line 11: | ||
*Comprehensive and quantitative analysis with LabChip GX software | *Comprehensive and quantitative analysis with LabChip GX software | ||
==[[BioMicroCenter:Caliper_INFO | Available Analyses]] == | |||
===HiSens DNA=== | |||
Sensitivity: | Sensitivity: | ||
| Line 24: | Line 24: | ||
*200 pg/μL for smears | *200 pg/μL for smears | ||
===Total RNA=== | |||
RNA metrics such as peak heights, peak areas, and concentration are used to determine an RQS (RNA Quality Score) number for each sample. The Caliper RQS is a calculated score that rates the quality of RNA samples. The RQS has been validated to correlate well with Agilent’s RIN (RNA Integrity Number) and follows the same 0-10 scale rating. Results comparing RIN to RQS for the same samples run on both LabChip GX and Agilent’s Bioanalyzer 2100 typically show <10% deviation. | RNA metrics such as peak heights, peak areas, and concentration are used to determine an RQS (RNA Quality Score) number for each sample. The Caliper RQS is a calculated score that rates the quality of RNA samples. The RQS has been validated to correlate well with Agilent’s RIN (RNA Integrity Number) and follows the same 0-10 scale rating. Results comparing RIN to RQS for the same samples run on both LabChip GX and Agilent’s Bioanalyzer 2100 typically show <10% deviation. | ||
== [[BioMicroCenter:Caliper_INFO | RNA Analyses]] == | |||
===How the Caliper Software Analyzes RNA Data === | |||
The LabChip GX RNA assay analysis determines the quality of the RNA sample by measuring the relative amounts of know RNA fragments relative to the total RNA present in the sample. Results for each well are calculated after all data for the well has been read. The data analysis process for RNA assays consists of the following steps: | The LabChip GX RNA assay analysis determines the quality of the RNA sample by measuring the relative amounts of know RNA fragments relative to the total RNA present in the sample. Results for each well are calculated after all data for the well has been read. The data analysis process for RNA assays consists of the following steps: | ||
| Line 61: | Line 60: | ||
===How to Read Caliper RNA Analysis Result === | |||
The Caliper RNA analysis results are almost the same as Agilent Bioanalyzer results. Caliper Life Sciences LabChip GX system analyzes RNA by electrophoretic separation on our microfluidic sipper chips. RNA molecules are separated and subsequently detected via laser induced fluorescence. The LabChip GX Software displays the raw data as an electropherogram and generates a gel-like image for visualization, data such as peak heights, peak areas, concentration, etc., are then calculated and stored as digitized data as text files and tables. These parameters are next used to determine an RQS (RNA Quality Score) number, a calculated score that rates the quality of RNA samples. | The Caliper RNA analysis results are almost the same as Agilent Bioanalyzer results. Caliper Life Sciences LabChip GX system analyzes RNA by electrophoretic separation on our microfluidic sipper chips. RNA molecules are separated and subsequently detected via laser induced fluorescence. The LabChip GX Software displays the raw data as an electropherogram and generates a gel-like image for visualization, data such as peak heights, peak areas, concentration, etc., are then calculated and stored as digitized data as text files and tables. These parameters are next used to determine an RQS (RNA Quality Score) number, a calculated score that rates the quality of RNA samples. | ||
| Line 85: | Line 84: | ||
=== Caliper RQS VS Agilent RIN === | |||
RNA metrics such as peak heights, peak areas, and concentration are used to determine an RQS (RNA Quality Score) number for each sample. The Caliper RQS is a calculated score that rates the quality of RNA samples. The RQS has been validated to correlate well with Agilent’s RIN (RNA Integrity Number) and follows the same 0-10 scale rating. Results comparing RIN to RQS for the same samples run on both LabChip GX and Agilent’s Bioanalyzer 2100 typically show <10% deviation. The RQS is consistent over Caliper’s standard sensitivity RNA assay concentration range (25 to 250 ng/μL) with CV’s <20%. For more information: http://www.caliperls.com/assets/022/8317.pdf | RNA metrics such as peak heights, peak areas, and concentration are used to determine an RQS (RNA Quality Score) number for each sample. The Caliper RQS is a calculated score that rates the quality of RNA samples. The RQS has been validated to correlate well with Agilent’s RIN (RNA Integrity Number) and follows the same 0-10 scale rating. Results comparing RIN to RQS for the same samples run on both LabChip GX and Agilent’s Bioanalyzer 2100 typically show <10% deviation. The RQS is consistent over Caliper’s standard sensitivity RNA assay concentration range (25 to 250 ng/μL) with CV’s <20%. For more information: http://www.caliperls.com/assets/022/8317.pdf | ||
== [[BioMicroCenter:Caliper_INFO | Software]] == | |||
The LabChip GX software for data analysis allows users to visualize results via an electropherogram or virtual gel view and provides data in tabular form, which can then be analyzed or easily exported into a spreadsheet format. The soft ware is available for download from BioMicro at http://biomicro-bioanalyzer.mit.edu/ | The LabChip GX software for data analysis allows users to visualize results via an electropherogram or virtual gel view and provides data in tabular form, which can then be analyzed or easily exported into a spreadsheet format. The soft ware is available for download from BioMicro at http://biomicro-bioanalyzer.mit.edu/ | ||
Revision as of 20:12, 18 March 2011
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |
CALIPER LAB CHIP

The Caliper LabChip GX is the most accurate and advanced nucleic acid separations system. The GX suite of instruments utilizes Caliper’s innovative microfluidics technology to perform reproducible, high-resolution, eletrophoretic separations.
- Complete assessment of RNA quality for better gene expression data
- Exact sizing and quantitation of DNA fragments
- Extreme accuracy with resolution down to 5 bp and sensitivity of 0.1ng/ul
- Flexible with 96 and 384 well compatibility
- Comprehensive and quantitative analysis with LabChip GX software
HiSens DNA
Sensitivity:
Standard Workflow
- 5 pg/μL per fragment
- 100 pg/μL for smears
Limited Sample Workflow
- 10 pg/μL per fragment
- 200 pg/μL for smears
Total RNA
RNA metrics such as peak heights, peak areas, and concentration are used to determine an RQS (RNA Quality Score) number for each sample. The Caliper RQS is a calculated score that rates the quality of RNA samples. The RQS has been validated to correlate well with Agilent’s RIN (RNA Integrity Number) and follows the same 0-10 scale rating. Results comparing RIN to RQS for the same samples run on both LabChip GX and Agilent’s Bioanalyzer 2100 typically show <10% deviation.
How the Caliper Software Analyzes RNA Data
The LabChip GX RNA assay analysis determines the quality of the RNA sample by measuring the relative amounts of know RNA fragments relative to the total RNA present in the sample. Results for each well are calculated after all data for the well has been read. The data analysis process for RNA assays consists of the following steps:
1. Raw data is read and stored by the system for each individual well.
2. The data is filtered and the resulting electropherograms of all wells are plotted. A curve spline fit to the data is performed to generate a baseline above which RNA fragment peaks are detected.
3. Peaks extending above the baseline are identified for all wells and are tabulated by migration time.
4. A sizing ladder (see the figure below), which is a mixture of RNA fragments of different known sizes, is run first from the ladder vial. The concentrations and sizes of the individual nucleotides present in the ladder are predetermined by the assay. A dye matching the lowest peak in the ladder is run with each of the samples. This lower marker, labeled LM in the RNA sample (see figure below) is used to align the ladder data with data from the sample wells.
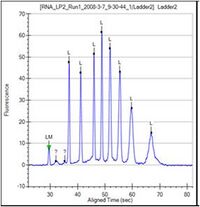
5. The ladder is analyzed and a standard curve of migration time versus RNA size is plotted from the RNA ladder by interpolation between individual RNA fragment size/migration points.
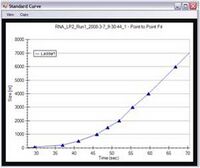
6. The standard curve and the markers are used to calculate RNA fragment sizes for each well from the migration times measured.
7. The Total RNA present is computed by finding the area under the electropherogram trace.The baseline for this integration is a straight line starting at the end of the lower marker and ending at the baseline end time. The height of the baseline endpoints is computed from an average of a five second window around the baseline start and end times.
8. The Total RNA concentration in the sample is computed from the ratio of the RNA area in the sample to the RNA area in the ladder multiplied by the ladder concentration specified in the assay.
9. Assay-defined RNA fragments are identified from the peaks in the peak table. Fragments are located by finding the largest peak within a size range associated with the fragment.For Eukaryote RNA assays, 5S, 18S and 28S fragments are located. For Prokaryote assays, 5S, 16S and 23S fragments are identified.

10. Messenger RNA Assay: The RNA contamination ratio is computed. This is the ratio of the area of all the fragments to total RNA area.
How to Read Caliper RNA Analysis Result
The Caliper RNA analysis results are almost the same as Agilent Bioanalyzer results. Caliper Life Sciences LabChip GX system analyzes RNA by electrophoretic separation on our microfluidic sipper chips. RNA molecules are separated and subsequently detected via laser induced fluorescence. The LabChip GX Software displays the raw data as an electropherogram and generates a gel-like image for visualization, data such as peak heights, peak areas, concentration, etc., are then calculated and stored as digitized data as text files and tables. These parameters are next used to determine an RQS (RNA Quality Score) number, a calculated score that rates the quality of RNA samples.
• Caliper RNA Ladder Result (Electropherogram of a typical RNA ladder): Total 9 peaks. The first peak is lower marker used to align the ladder data with data from the sample wells. Others represent different sizing ( 200nt, 500nt, 1000nt, 1500nt, 2000nt, 3000nt, 4000nt and 6000nt).

• Caliper RNA Eukaryote Sample Result The electropherogram for typical total RNA samples is shown below. Your results may vary depending on the type of total RNA.

• Caliper RNA Prokaryot Sample Result The electropherogram for typical Prokaryote Total RNA samples is shown below. Your results may vary depending on the type of total RNA.

• RNA mRNA Sample Result The electropherogram for a typical mRNA sample is shown below. Your results may vary depending on the type and concentration of mRNA.

Caliper RQS VS Agilent RIN
RNA metrics such as peak heights, peak areas, and concentration are used to determine an RQS (RNA Quality Score) number for each sample. The Caliper RQS is a calculated score that rates the quality of RNA samples. The RQS has been validated to correlate well with Agilent’s RIN (RNA Integrity Number) and follows the same 0-10 scale rating. Results comparing RIN to RQS for the same samples run on both LabChip GX and Agilent’s Bioanalyzer 2100 typically show <10% deviation. The RQS is consistent over Caliper’s standard sensitivity RNA assay concentration range (25 to 250 ng/μL) with CV’s <20%. For more information: http://www.caliperls.com/assets/022/8317.pdf
The LabChip GX software for data analysis allows users to visualize results via an electropherogram or virtual gel view and provides data in tabular form, which can then be analyzed or easily exported into a spreadsheet format. The soft ware is available for download from BioMicro at http://biomicro-bioanalyzer.mit.edu/
